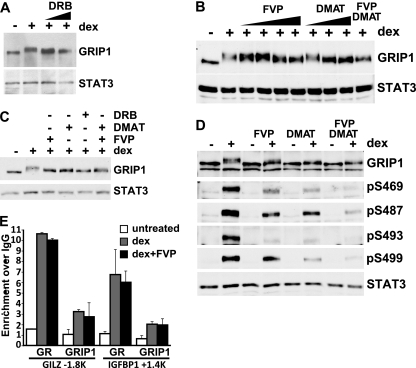
Fig 5.
CK2 and CDK9 target a subset of MS-identified GRIP1 phosphorylation sites in vivo. (A) U2OS-rGR cells were untreated or treated for 2 h with 100 nM dex with or without DRB (50 or 100 μM), and GRIP1 and STAT3 were detected by immunoblotting. (B) U2OS-rGR cells were untreated or treated for 2 h with 100 nM dex with or without FVP or DMAT in increasing concentrations (5, 10, 50, and 100 nM and 1, 5, and 10 μM, respectively) or 100 nM FVP plus 10 μM DMAT together, and GRIP1 and STAT3 were detected by immunoblotting. (C) U2OS-rGR cells were untreated or pretreated with 100 nM FVP, 10 μM DMAT, 200 μM DRB, or FVP plus DMAT together, as indicated, for 2 h, followed by the addition of 100 nM dex for an additional 2 h, and GRIP1 and STAT3 were detected by immunoblotting. (D) U2OS-rGR cells were incubated for 2 h with (+) or without (−) 100 nM dex in the absence or presence of FVP, DMAT, or FVP plus DMAT (following pretreatment with inhibitors for 45 min), as indicated, and immunoblotting was performed with antibodies to GRIP1, antisera to indicated GRIP1 phosphosites, and STAT3. (E) U2OS-rGR cells were untreated or pretreated for 30 min with 100 nM FVP, followed by the addition of 100 nM dex for 30 min, and ChIPs were performed with antibodies to GR (35), GRIP1 (Bethyl Laboratories), or normal rabbit IgG as negative control. Genomic regions corresponding to the GILZ and IGFBP-1 GREs at approximately kb −1.8 and +1.4, respectively, were amplified and normalized to corresponding signals at the unrelated 28S rRNA gene, and the results are expressed relative to the signal obtained with normal IgG (set to 1). Error bars show standard errors of the mean.