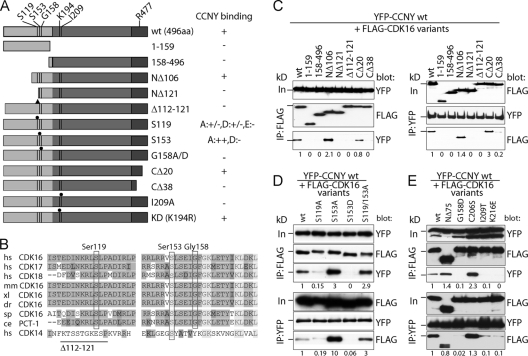
Fig 2.
Mapping of CDK16 residues required for CCNY binding. (A) Scheme of N-terminally FLAG-tagged CDK16 variants analyzed for interaction with YFP-tagged CCNY. (B) Amino acid sequences of ∼60 residues upstream of the conserved kinase domain were aligned with each other. Identical residues are shown in medium gray and similar ones in dark gray, whereas completely conserved residues are shown in light gray. The positions of human Ser119, Ser153, and Gly158 as well as the deletion of residues 110 to 121 are indicated. hs, human; mm, mouse; xl, frog; dr, zebrafish; sp, sea urchin; ce, nematode. (C) 293A cells were transfected with expression plasmids for YFP-CCNY together with FLAG-tagged CDK16 variants for 60 h. IPs were performed as in Fig. 1, and proteins were immunoblotted for the presence of the interaction partner. (D and E) Cells were transfected as in panel C to express phosphosite mutants of FLAG-CDK16 and tested for YFP-CCNY binding. Input (In) is 10% of the protein amount used for IP. The position of the 72-kDa protein marker band is indicated. Proteins were quantified by scanning densitometry using ImageJ. Relative levels normalized to the values of the immunoprecipitated partner proteins are shown.