Abstract
Basal bodies and centrioles play central roles in microtubule (MT)-organizing centres within many eukaryotes. They share a barrel-shaped cylindrical structure composed of nine MT triplet blades. Here, we report the structure of the basal body triplet at 33 Å resolution obtained by electron cryo-tomography and 3D subtomogram averaging. By fitting the atomic structure of tubulin into the EM density, we built a pseudo-atomic model of the tubulin protofilaments at the core of the triplet. The 3D density map reveals additional densities that represent non-tubulin proteins attached to the triplet, including a large inner circular structure in the basal body lumen, which functions as a scaffold to stabilize the entire basal body barrel. We found clear longitudinal structural variations along the basal body, suggesting a sequential and coordinated assembly mechanism. We propose a model in which δ-tubulin and other components participate in the assembly of the basal body.
Keywords: basal body, centriole, Chlamydomonas reinhardtii , electron cryo-tomography, three-dimensional reconstruction
Introduction
The basal body, as a cellular organelle for organizing microtubules (MTs), is composed of nine MT triplet blades. It carries out multiple essential functions in cellular processes. During cell division, it functions as a centriole to recruit pericentriolar material to form the centrosome, which in turn is responsible for establishing the bipolar spindle during mitosis. Although centrioles are dispensable for mitosis in certain organisms and cell lines (Debec et al, 2010), ample evidence shows that in many other cases, over-amplification or depletion of centrioles leads to a delay in cell division, aneuploidy and cell death, suggesting their importance for normal mitosis (Hinchcliffe et al, 2001; Piel et al, 2001; Mikule et al, 2007). More importantly, the centriole is responsible for establishing cell polarity during cell division (Yamashita et al, 2007; Wang et al, 2009). In many quiescent cells, the centriole will migrate to the cell periphery, and anchor beneath the cell membrane as a basal body that templates cilium formation. The basal body also provides a docking site for intraflagellar transport particles that move bidirectionally along the cilium (Deane et al, 2001). Recent studies show that, in addition to its role in motility, the cilium also functions as an antenna for communicating between the intracellular and external environment of the cell and plays essential roles during cell regulation, tissue differentiation and embryonic development (Nigg and Raff, 2009). Mutations in cilium or basal body genes result in various forms of human disease, collectively known as ciliopathies (Gerdes et al, 2009).
Biogenesis of the centriole and basal body has been studied in a variety of model organisms (Ringo, 1967; Dippell, 1968; Allen, 1969; Anderson and Brenner, 1971; Anderson, 1972; Cavalier-Smith, 1974; González et al, 1998; Pelletier et al, 2006; Giddings et al, 2010; Guichard et al, 2010). Genes involved in centriole biogenesis, either regulatory or structural, are generally well conserved, suggesting a common assembly pathway across species (Carvalho-Santos et al, 2010). The biogenesis of basal bodies or centrioles is tightly coupled to the cell cycle, suggesting a highly regulated assembly mechanism (Doxsey et al, 2005).
Proteomic studies of purified basal bodies or centrioles have identified a list of bona fide centriole components and basal body specific proteins many of whose function and structure remain unknown (Andersen et al, 2003; Li et al, 2004; Keller et al, 2005; Kilburn et al, 2007). Despite extensive descriptions of the centriole and basal body ultrastructure, the lack of high-resolution structural information significantly limits our understanding of the assembly process and the full repertoire of cellular functions for this complex organelle.
Here, we used electron cryo-tomography (cryo-ET) to visualize basal bodies purified from Chlamydomonas reinhardtii in a near-native state. In combination with a subtomogram averaging strategy, it has been possible to obtain a high-quality 3D reconstruction of the basal body that offers insight on how tubulin protofilaments (PFs) and accessory components assemble into the triplet. Based on our structural findings and previous studies, we propose potential locations for several basal body components. Our structure provides a framework for understanding the molecular mechanism of basal body and centriole biogenesis, and for integrating new information as the protein composition of the non-MT components are elucidated.
Results
Overall structure of the basal body triplet
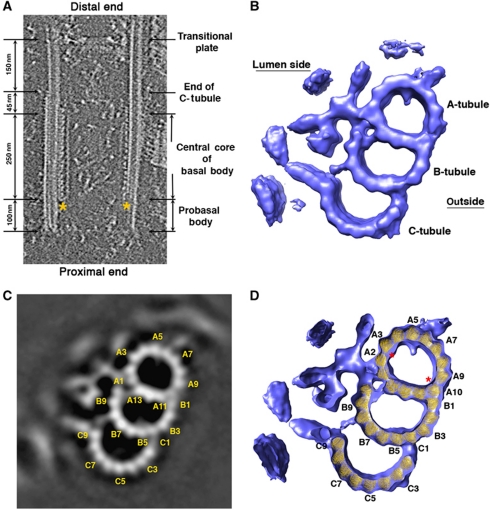
Purified basal bodies in near-native state were visualized by cryo-ET. In most of the tomograms, two cylindrically shaped basal bodies were visible since they are tethered laterally by the distal striated fibres (Hoops et al, 1984). The basal bodies have a reproducible size, with an average diameter of 260 nm and an overall length of 600 nm (Figure 1A). The MT triplets start from the proximal end (the bottom in Figure 1A) and span about 400 nm longitudinally towards the distal end where the C-tubules of the triplets terminate. The A- and B-tubules continue as a doublet for about 150 nm before they reach the transitional plate, a hallmark of the transition zone where the axoneme will assemble (Cavalier-Smith, 1974; O’Toole et al, 2003; Geimer and Melkonian, 2004). We did not observe the cartwheel structures in our tomograms, presumably because they were lost during purification. Electron dense structures, known as ‘A-tubule feet’, are consistently visible in our tomograms along the wall of the triplets projecting towards the lumen (Cavalier-Smith, 1974; Geimer and Melkonian, 2004), showing characteristic 8- and 16-nm periodicity (Figure 1A). The region decorated with A-tubule feet starts at about 100 nm from the triplet minus (proximal) end, spans about 250 nm longitudinally and terminates at about 45 nm before the triplets become doublets. This section forms the central core of the basal body and was used for tomogram subvolume averaging in order to obtain a higher-resolution structure of the basal body.
Figure 1.
Cryo-ET reconstruction of the basal body triplet. (A) A cross-section of tomographic reconstructed volume containing a basal body. The section is through the centre of the basal body barrel. The proximal end is at the bottom and the distal end is at the top. The ‘A-tubule feet’ are marked with *. The terms of probasal body, A-tubule feet and transitional plate follow conventions of Geimer and Melkonian (2004). (B) The averaged 3D structure of the basal body triplet viewed from the distal end. The MT triplet density map has been deposited in the Electron Microscopy Data Bank with accession code EMD-5252. (C) A longitudinal projection of a 16-nm section of the averaged triplet. Every other PFs in the triplet are labelled. (D) Docking of crystal structure of tubulin into the triplet density map. Selected PFs are labelled. The locations of the highest curvature in the A-tubule are marked with *.
We obtained an averaged MT triplet at 33 Å resolution (Figure 1B; Supplementary Figure S1). The resolution of the averaged structure is sufficiently high to allow us to discern each PF in the longitudinal projection of the triplet (Figure 1B and C). The A-tubule is a complete MT composed of 13 PFs, numbering starts clockwise from the basal body luminal side as A1 to A13 following the Tilney-Linck convention (Linck and Stephens, 2007). PF A10 is the site where the B-tubule joins with the A-tubule. PFs A10 to A13 are referred to as partition PFs shared between A- and B-tubule. The B-tubule is composed of 10 PFs that are numbered from the outside surface of the triplet clockwise as B1 to B10. The C-tubule also has 10 PFs. PF C1 starts as a branch from PF B4 and they are numbered clockwise as C1 to C10. PFs B5 to B8 form the partition shared by the B- and C-tubules.
The A-tubule has an elliptical shape with a substantial variation of curvature along the MT wall (Figure 1B and C). The longest axis running across the internal diameter of the A-tubule is between PFs A3 and A10 with a diameter of ∼247 Å. The shortest axis is ∼201 Å between PFs A6 and A13. This change of diameters is equivalent to a 10% distortion of an intact 13-PF MT and is similar to the A-tubule in the axoneme doublet where ∼8% distortion has been observed (Sui and Downing, 2006). Interestingly, the variation of curvature is non-isotropic along the wall of the A-tubule. The highest curvature is at PFs A9 and A10, followed by the next-highest curvature at PFs A2 and A3, where large lateral gaps are observed between PFs. The smallest curvature is at the partition site from A11 to A13. In contrast to the A-tubule, the B- and C-tubules have a rather smooth and uniform curvature with diameters about 260 Å. The diameter and the curvature of the B- and C-tubules indicate that both would form a circular MT with 15 PFs if they formed complete rings, similar to the B-tubule in the axoneme doublet (Sui and Downing, 2006).
Building a pseudo-atomic model of the triplet
In our structure, the 4-nm periodicity between individual tubulin monomers can be easily resolved along most of the PFs of the basal body (Supplementary Figure S2). The z-rise (longitudinal rise) of tubulins between adjacent PFs, varies from 10 to 12 Å, consistent with previous theoretical and experimental data from MTs with different PF numbers (Chrétien and Wade, 1991; Sui and Downing, 2010). This allowed us to fit the atomic structure of α/β tubulin into the EM density map and build a pseudo-atomic model of the tubulin core of the triplet (Figure 1D). Previous studies have shown that in α/β tubulin, the M-loop and H1-S2/H2-S3 loops provide the main lateral interactions between adjacent PFs and we have kept these contacts in our model (Nogales et al, 1999; Li et al, 2002; Sui and Downing, 2010). The overall fit is excellent at the current resolution. In the A-tubule, due to the variation in local curvature, the closest lateral interactions are within partition PFs A11 to A13. Conversely, the interfaces between PFs A2/A3 and PFs A9/A10 have less contact due to large local curvature.
The fitting of tubulin PFs into the B- and C-tubule density resulted in uniform tubulin lateral interactions. We have modelled PF B1 as a tubulin PF that makes unusual lateral contact with the outside surface of PF A10. Since the distal half of PF C1 exhibits an 8-nm interval with a gap (Figure 4A and B), it is most likely occupied by non-tubulin protein in this region. Therefore, we did not fit tubulin monomers into the PF C1 position anywhere along its length. This longitudinal change in PF C1 will be described in detail below.
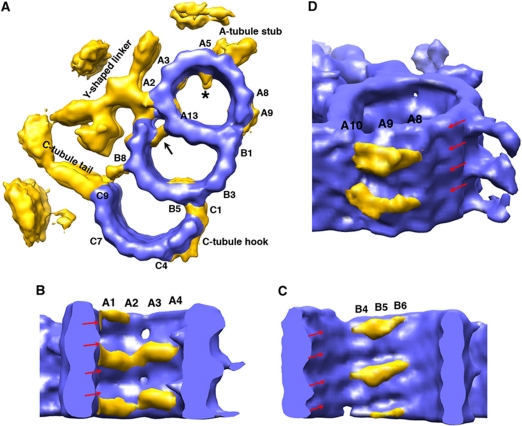
Figure 4.
Longitudinal structural variations observed in the triplets. (A) Subvolumes are divided into eight groups and averaged according to their longitudinal position in the basal body. The relative positions of the group are highlighted with the yellow boxes. Each group contains a triplet segment about 39 nm in length. Structural differences are shown with cross-sectional views of the C-tubule for each averaged triplet, cutting through PFs C3/C7, PFs B4/C1/C2, respectively. The scale bar is 10 nm. (B) Comparison of the two triplet structures resulted from averaging of the proximal half (in green) and the distal half (in blue) separately. The hook-like linkers that connect the B- and C-tubule in the distal half triplet are coloured in yellow. The arrows indicate the positions of C1. (C) Filaments (in yellow) are observed in the lumen of the C-tubule in the distal half of the triplet, with a longitudinal spacing every 8 nm. The tubulin helices are indicated with red arrows. The longitudinal length of the triplets in Figure 4B and C is about 26 nm.
Non-tubulin components associated with the triplet
Proteomic studies have shown that, besides tubulin, the basal body contains nearly 50 non-tubulin components (Keller et al, 2005). In order to find where they bind, how the tubules are associated to form a triplet and how the triplets are connected in the basal body, we used the pseudo-atomic model of the tubulins within the triplet as a mask to subtract its density from the 3D density map. The resulting difference map shows novel density features that must correspond to the locations of most of the non-tubulin accessory proteins (Figure 2A).
Figure 2.
Non-tubulin densities associated with the basal body triplet. (A) A calculated difference map based on the pseudo-atomic model of the triplet. The density contributed by tubulin is coloured in blue and the non-tubulin proteins associated with the triplet are coloured in yellow. Selected PFs are labelled. The arrow points to the second A- and B-tubule linker connecting PFs A13 and B10. A cone-like structure in the lumen of the A-tubule is marked with *. (B) Filamentous structure associated in the luminal wall of the A-tubule. The filaments attached to the luminal side of A-tubule are highlighted in yellow. They are running across PFs A1 to A4, with 8-nm periodicity. The tubulin helices in the A-tubule are indicated with red arrows. (C) Filamentous structure associated on the luminal wall of the B-tubule. The densities are running across PFs B4 to B6, with 8-nm periodicity. The tubulin helices in the B-tubule are indicated with red arrows. (D) Densities are observed on the outside wall of the A-tubule, running across PFs A8 to A10 with a longitudinal spacing of every 8 nm. The red arrows indicate tubulin helices in the A-tubule.
Just as in the axoneme doublet structure (Nicastro et al, 2006; Sui and Downing, 2006), we also observed densities decorating on the internal wall of A- and B-tubules in the triplet, but in a more asymmetric manner. In the A- and B-tubule, there are filamentous densities running across PFs A1 to A4 and B4 to B6, respectively, with an 8-nm periodicity (Figure 2B and C). The densities follow the rise of the tubulin helical repeat inside the lumen, forming lateral cross-links between neighbouring tubulin monomers at the lumen side. Likely, these are Tektin family proteins that stabilize adjacent PFs and fine-tune local curvature of the tubule (Amos, 2008). We also observed a cone-shaped density attached on the luminal side of PF A5 with 8-nm periodicity (* in Figure 2A), likely to stabilize the A-tubule or as the part of structure connecting to the neighbouring triplet. Interestingly, similar density has been consistently observed in the axoneme doublet (Nicastro et al, 2006; Sui and Downing, 2006; Movassagh et al, 2010). Across from the cone-shaped density, at PF A6 a stub-like density projects out every 8 nm on the outside wall of the A-tubule (Figure 2A). The stub connects to the C-tubule from the neighbouring triplet and will be described in detail in the next section. Meanwhile, density was observed on the outside wall of the A-tubule running across PFs A8 to A10 with an 8-nm longitudinal spacing (Figure 2D). Since both PFs A10 and B1 are at the junction of A- and B-tubules, this density presumably will stabilize the linkage between the A- and B-tubules at the outside wall of triplet.
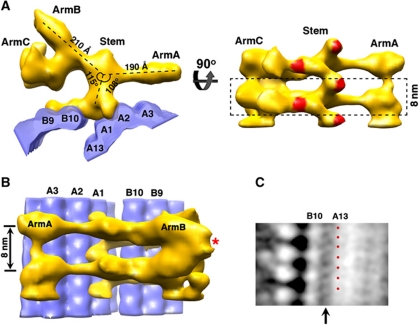
Remarkably, a large Y-shaped density was observed lying horizontally as a ridge along the luminal side of the A- and B-tubules (Figures 2A and 3A). It spans about 380 Å, nearly the entire inner circumference of the A- and B-tubules. It projects radially about 170 Å towards the centre of the basal body barrel. The estimated mass of this structure is 1.1 MDa. We have divided this structure into four parts and assigned them as a central stem with three arms, termed armA, armB and armC (Figure 3A and B). The central stem binds directly to both A- and B-tubule. On the right side in Figure 3A, it fits into the groove between PFs A1 and A2, which is likely the position of the seam in the A-tubule (Song and Mandelkow, 1995). After rising longitudinally ∼40 Å, the left side of the stem binds to PF B10 in the B-tubule. Here, the stem extends laterally, running across the entire outer surface of PF B10 and partial surface of PF B9. Together, this central stem fills in the gap between the A- and B-tubule and cross-links the two tubules at their closest distance (∼82 Å). It makes substantial interactions with both tubules at their luminal joint, with an estimated total contact area between the stem and the A- and B-tubule of ∼5200 Å2. Multiple copies of the stem stack longitudinally with an axial repeat of 8 nm, forming a left-handed spiral-shaped filament viewed from the outside of the basal body (Figure 3A). This filament might account for the 11th PF of the B-tubule as previously observed (Tilney et al, 1973).
Figure 3.
Link between the A- and B-tubules in the basal body triplet. (A) Two views showing a large Y-shaped structure lying horizontal between the A- and B-tubules at the luminal side of the basal body. A stack of these structures forms a longitudinal filament assembly. The model of MT PFs is in blue. A dashed-line box shows a single 8-nm longitudinal repeat unit. The areas that contact the A- and B-tubules are highlighted in red. (B) The Y-shaped structure viewed from the luminal side of the basal body. The model of MT PFs is in blue. Two neighbouring armBs join and form a bifurcated structure as indicated with *. (C) A second A- and B-tubule linker connecting PFs A13 and B10 as indicated with an arrow. The 4-nm periodicity is highlighted with red dots.
ArmA rotates about 108° counterclockwise relative to the stem and extends 190 Å towards the direction of A-tubule (Figure 3A and B). At its end, armA connects longitudinally to the neighbouring armAs, resulting in a filament along the basal body barrel with an 8-nm periodicity. ArmB is 210 Å long and it is about 115° clockwise relative to the stem (Figure 3A). At the end of armB, the neighbouring two arms join, forming in a bifurcated structure with 16-nm periodicity (Figure 3B). This contributes to the 16-nm axial repeat pattern projecting towards the lumen of the basal body barrel as observed in the tomograms (Figure 1A). ArmC is branched 90° from the armB, with a hook-shaped structure. Like armA, a stack of armC also connects longitudinally to form a filament with 8-nm periodicity (Figure 3A). In conclusion, this large Y-shaped structure forms a scaffold on the luminal side of the basal body cross-linking and probably stabilizing both A- and B-tubule.
In addition to the large density that connects the A- and B-tubules as described above, we also observed relative weak filamentous densities that connect PFs B10 and A13 on the inside of the tubule structure (Figures 2A and 3C). The connection, which spans about 90 Å, forms a ladder with 4-nm periodicity. Each rung of the ladder is tilted, with a rise about 4 nm from one end at PF B10 to the other end at PF A13, further stabilizing the linkage between A- and B-tubule.
Longitudinal structural variations along the basal body
To reveal the structural variations of the basal body, we have divided the triplet subvolumes into eight groups according to their longitudinal positions. Only subvolumes within each group were averaged (Figure 4A). To our surprise, groups from the proximal half of basal body (groups 1–4) have distinct structural variations compared with the groups from the distal half (groups 5–8). The variations mainly reside on the C-tubule. From group 4 to group 5, there is an emergence of density in the lumen of the C-tubule. In addition, the periodicity at PF C1 changes from 4 to 8 nm. To analyse these changes in greater detail, we re-divided the subvolumes into two groups, the proximal half (sum of 972 subvolumes from groups 1–4) and the distal half (sum of 972 subvolumes from groups 5–8) and recalculated their averaged structure, respectively. The resulting two EM density maps, as shown in Figure 4B, show significant differences. In the proximal half, the C1 PF, like any other MT PFs in the C-tubule, exhibits a 4-nm axial repeat. In contrast, the distal half has longitudinally disconnected density with 8-nm periodicity. In addition, the density with 8-nm spacing at C1 extends further laterally across the outside surface of PF C2, C3 and C4, forming a continuous hook-shaped arm with an estimated mass of 270 kDa. This arm forms a stable linker between the B- and C-tubules at the outside surface of the basal body.
The distal half of the basal body additionally contains a stack of crescent-shaped filamentous structures (Figure 4C). These filaments span ∼136 Å in the lumen of the C-tubule. One end of the filament anchors to PF C3, while the other end attaches to the junction between PFs C6 and C7. The estimated mass of this elongated filament is about 46 kDa. Interestingly, the filament is tilted and descends about 20 Å running from PF C3 to PFs C6/C7. Assuming the triplet C-tubule has the MT B-surface lattice and can be seen as a bundle of tubulin helices (red arrows in Figure 4C) (Chrétien and Wade, 1991; Song and Mandelkow, 1995), the tilting of the filament is in the opposite direction to the tilting of the tubulin helices, indicating that the luminal filament most likely attaches to one tubulin helix at the C3 end and attaches to the adjacent tubulin helix running underneath at the C6/C7 end. These attachments suggest that the filament has intrinsic polarity with two ends making unique contacts with the C-tubule. The luminal densities in the MT have been reported previously and presumably stabilize the local MT structure (Garvalov et al, 2006; Cyrklaff et al, 2007). The filamentous structure that we describe in the lumen of the C-tubule could function as a reinforcement beam to enhance C-tubule rigidity in the distal half of the basal body.
The emergence of the luminal filaments at the distal half of the basal body coincides with a structural change at PF C1. These transitions take place near the middle of the basal body, about 225 nm from the triplet minus (proximal) end (Figure 4A). The two structures also overlap spatially as the hook-shaped arm runs across PFs C1 to C4 and the luminal filament spans PF C3 and PF C6/C7. It is conceivable that they might interact directly to participate in coordinated assembly of the basal body C-tubule.
Building a basal body model
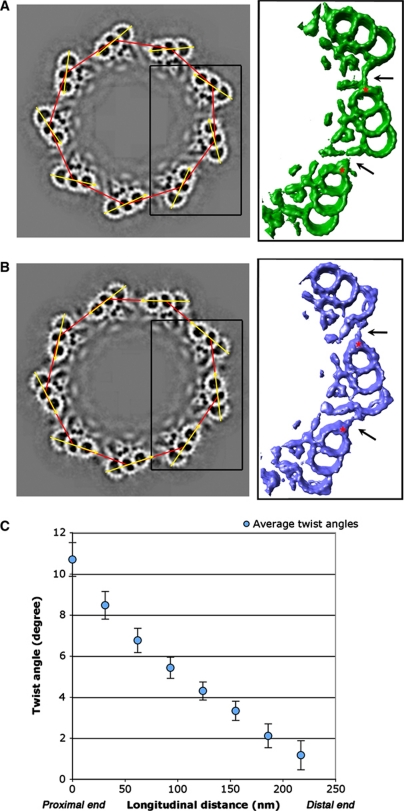
Most basal bodies and centrioles are composed of nine triplets interconnected circumferentially to make a barrel-shaped structure. In order to find out how the triplets are linked, we reconstructed the entire basal body based on the averaged triplet structure. Since the basal bodies were visualized in their near-native state and subvolumes containing triplet segments were computationally extracted from the tomogram for alignment and averaging, their spatial coordinates in the context of the entire basal body volume are known. By using the known subvolume coordinates, we replaced the subvolumes in each basal body with the averaged triplet structures (as shown in Figure 4B) and reconstructed entire longitudinal segments of the basal body without imposing nine-fold symmetry. The results as shown in Figure 5A and B represent the basal body proximal and distal ends, respectively. To our surprise, a comparison of the two ends reveals that the inter-triplet linkages are significantly different (black arrows in the right panel of Figure 5A and B, respectively). At the proximal end, a pointed stub-like density projects from PF A6 and connects to the neighbouring triplet at the end of the extended tail of the C-tubule. This inter-triplet linker is about 130 Å long from the base of the stub to the tip of the extended tail. In contrast, at the distal end, the A6 stub connects directly to the outside wall of the neighbouring C-tubule at PFs C7 and C8 with an approximate length of 170 Å.
Figure 5.
Building the basal body model. (A, B) Cross-section views of the reconstructed basal body at the proximal end (A) and at the distal end (B). Depending on the longitudinal position in the basal body, the proximal or the distal half averaged triplet structure (as shown in Figure 4B) is used to generate the basal body model. A nonagon is formed by connecting nine triplets with the red lines. The triplet orientations are indicated with the yellow arrows. The angle between each yellow arrow and its nearest red line was measured for each triplet. The right-side panel shows enlarged views of the boxed area in the basal body. The black arrows indicate the inter-triplet linkers. The locations of the PF A6 are highlighted with *. (C) The longitudinal twisting of the triplet in the basal body. The horizontal axis represents the relative longitudinal distance from the proximal to the distal end of the basal bodies. Each basal body was divided longitudinally into eight sections, as shown in Figure 4A. In each section, the twist angles from nine triplets, as defined in the main text, were measured. Total eight basal bodies were used for the measurement, resulting in 72 angle measurements at each longitudinal position of the basal body. The triplet twist angles were then plotted against the longitudinal distance. The error bars indicate the 95% confidence intervals.
To further analyse the change of this inter-triplet linkage, we measured the angle between two adjacent triplets. As shown in Figure 5A and B, a nonagon is created by connecting nine triplet centres in the basal body (red lines), in this case the centres of B-tubule. Meanwhile, a vector (yellow arrow) can be drawn across PFs A6–B6–C5/C6, indicating orientation for each triplet. The angle between the triplet vector and the nearest side of nonagon defines the relative angle of the triplet. Eight basal bodies were used for the angle measurement and they are summarized in Figure 5C. At the proximal end of the basal body, the average angle between the triplet vector and the nearest side of nonagon is 10.9°. However, this angle decreases gradually and reaches 1.0° at the distal end of the basal body. The gradual change of the angle indicates that each triplet twists about its centre at 0.04°/nm in a left-handed fashion from the proximal end to the distal end. The diameter of the basal body barrel and the relative location of the triplets remain constant in our models. This observation is consistent with previous studies on mammalian centrioles, where a twist angle of up to 25° has been observed and the diameter of the basal body barrel remained constant in the absence of EDTA (Anderson, 1972; Paintrand et al, 1992). This longitudinal left-handed twist of the triplet along the basal body results in a structural change at the inter-triplet linker. Since this twist proceeds gradually, it suggests that the inter-triplet linker is flexible. Alternatively, there might be multiple attachment sites for the inter-triplet linker running across the wall of the C-tubule. The mechanism for this transition awaits molecular details achieved only from higher-resolution structures.
Discussion
Comparison with the axoneme doublet
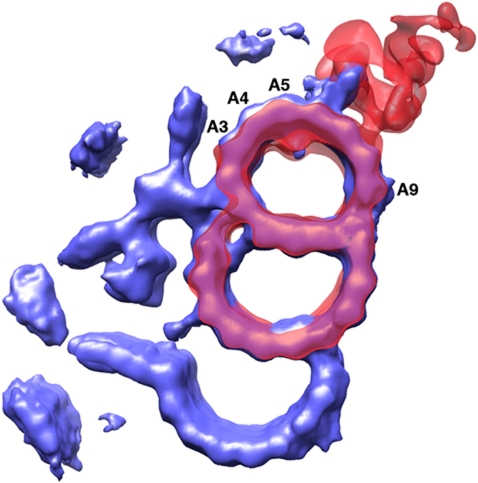
The basal body provides a template for axoneme assembly. To compare the two structures, we superimposed the triplet EM density with one of the available axoneme doublet structures at a comparable resolution (EMD-1696) (Movassagh et al, 2010). As shown in Figure 6, the overall fitting of the two structures is excellent. However, there are notable differences in the shape of the A-tubule and striking differences in non-MT densities. In both structures, the A-tubules are elliptically deformed. The A-tubule ring in the doublet is radially elongated ∼8% where the long axis of the elliptical ring runs across PFs A2/3 and A9. The distortion is likely due to accessory proteins, such as Tektins, attached to the luminal wall of the A-tubule (Sui and Downing, 2006). In the triplet, the A-tubule ring is elongated ∼10% in the same direction as the doublet. However, the PF A3, A4, A5 are stretched further outwards compared with the doublet (Figure 6). This results in an asymmetric elliptical shape of the A-tubule in the triplet. The variation of the local curvature demonstrates the intrinsic property of MT with flexible lateral PF contacts. This flexibility is consistent with the results from the EM reconstruction of in vitro assembled MTs with various PF numbers (Sui and Downing, 2010). Interestingly, the association patterns of the accessory proteins are also different in these two structures. In the doublet, substantial density has been observed in the lumen of the A-tubule and on both sides of the partition bridge, presumably to stabilize the doublet in resistance to the bending force generated by axonemal dynein (Nicastro et al, 2006; Sui and Downing, 2006). In contrast, the triplet has fewer accessory proteins attached to the luminal wall of A- and B-tubule, likely because the basal body bears less mechanical stress from its cellular environment and the A- and B-tubule are further bolstered by the C-tubule.
Figure 6.
Comparing the surface-rendered map of the basal body triplet with the previously published map of the axoneme doublet. The basal body triplet is coloured in blue. The doublet map (EMD-1696) is coloured in red (Movassagh et al, 2010).
Luminal structure cross-linking the A- and B-tubules in the triplet
One of the striking features presented in our structure is the horizontal Y-shaped structure circumferentially bound to the triplet on the luminal side of the basal body. This is significantly different from the axoneme doublet, where a flexible linker density with 16-nm periodicity connects the A- and B-tubules (Sui and Downing, 2006). In the triplet, the A- and B-tubules are tightly linked by a spiral-shaped filament formed by the stems that contact the A- and B-tubules alternately with a 4-nm rise (Figure 3A and B). In addition, the structure also has three extended arms emanating from the stem. Both armA and armC form longitudinal filaments with an 8-nm periodicity, while two neighbouring armBs join to form a bifurcated structure with a 16-nm repeat. Furthermore, in our basal body models, nine of these structures nearly fill the entire inner circumference of the basal body (Figure 5A and B). It is likely that they are connected by flexible linkers and form a cylindrical structure as observed previously (Geimer and Melkonian, 2004). In additional to the cartwheel and other basal body components, this might provide another scaffold inside the barrel of the basal body. Several functions might be carried out by this scaffold structure. First, it links the A- and B-tubules and reinforces the triplets. Second, it provides another inter-triplet linker besides the link between neighbouring A- and C-tubules. Third, the longitudinal interactions in these filaments will further stabilize the basal body barrel. Finally, this scaffold structure might recruit other luminal components, such as filaments and electron dense materials often seen in the central luminal region of the basal body (Cavalier-Smith, 1974; Geimer and Melkonian, 2004).
Similar luminal structures have previously been observed in centrioles and basal bodies. For example, luminal disks were observed in human centrioles (Paintrand et al, 1992; Ibrahim et al, 2009), and luminal rims were observed in the centrioles and basal bodies in other vertebrate cells (Fais et al, 1986). Although their detailed structures might be different from what we presented here, it is likely that luminal scaffold is a common feature of basal bodies and centrioles in many organisms, presumably playing a role both in stabilizing the assembly and in recruiting additional components. This luminal scaffold emerges as a rigid structure about 100 nm from the minus (proximal) end of the basal body and spans about 250 nm longitudinally (Figure 1A). This is concomitant with the transition of the probasal body to the basal body, suggesting that it assembles during elongation and early maturation of the basal body in G2 phase in Chlamydomonas (Piasecki et al, 2008). However, in mammalian cells, this might take place in early S phase when the procentrioles start to elongate (Vorobjev and Chentsov, 1982).
Longitudinal variations and its implications for the basal body assembly
Perhaps the most remarkable finding from our structure is the structural changes between the proximal and the distal half of the basal body. These changes are mainly confined to the C-tubule, but effectively alter assembly of the whole basal body. One of the Chlamydomonas genes that affect the C-tubule formation is the UNI3 gene encoding δ-tubulin (Dutcher and Trabuco, 1998). Previous studies have shown that deletion of δ-tubulin led to a basal body with mostly doublets and occasional short stretches of triplet at the distal end, suggesting that the δ-tubulin is required for extension and stability of the C-tubule (Garreau de Loubresse et al, 2001; Fromherz et al, 2004). Additional defects of the δ-tubulin deletion include duplicated and misplaced transition zones and a misaligned cell cleavage furrow (O’Toole et al, 2003). Since one of the major structural changes we have observed is at PF C1, it strongly suggests the PF C1 is composed of δ-tubulin. Comparing to α/β tubulins, δ-tubulin has a unique and highly conserved insertion sequence near the M-loop located at the tubulin lateral interface. This insertion will be ideally suited to make an unusual lateral contact with PF B4 from the B-tubule (Chang and Stearns, 2000; Inclán and Nogales, 2001). Interestingly, an α-tubulin mutant encoded by TUA2 could suppress the δ-tubulin deletion phenotype (Fromherz et al, 2004). This α-tubulin mutant might partially replace δ-tubulin at the proximal half of PF C1 and partly restore triplet formation. As the assembly extends further towards the middle of the basal body, δ-tubulin will be replaced by an unknown protein or complex with a mass of about 270 kDa. It binds to PF B4 every 8 nm and laterally cross-links PF C2, C3 and C4. This complex extends longitudinally along the PF C1 as a filament until the C-tubule terminates. In the absence of δ-tubulin, this complex may only partially bind to PF B4 and assembles as PF C1, resulting in short stretches of triplet as observed at the distal end of the basal body in the δ-tubulin deletion mutant (O’Toole et al, 2003), further supporting the notion that the extension of PF C1 and C-tubule is a cooperative assembly process.
In contrast to δ-tubulin, a conserved centriole protein POC5 is localized mainly at the distal portion of the centriole and is essential for procentriole elongation to its full length (Azimzadeh et al, 2009). Like δ-tubulin, depletion of POC5 resulted in centrioles with MT doublets, suggesting that POC5 might be involved in the extension of C-tubule during the centriole assembly. Interestingly, both δ-tubulin and POC5 genes are absent in organisms such as Caenorhabditis elegans and Drosophila melanogaster, where only singlets or doublets are observed in the centriole or basal body in the somatic cells (Dutcher, 2001; Azimzadeh et al, 2009), further suggesting their unique roles in assembly of the C-tubule of the triplet. However, triplets have been observed in the basal bodies and centrioles in germ cells in Drosophila (Mahowald and Strassheim, 1970; Riparbelli and Callaini, 2011). It will be interesting to investigate the tissue-specific expression levels of different tubulin isoforms, as this might provide insight into their specific roles during the basal body/centriole assembly.
Detailed analysis of POC5 also revealed that procentriole elongation consists of two sequential and distinct steps coupled to cell-cycle progression (Azimzadeh et al, 2009). The structural variations observed in our study of the basal body, in which the proximal half exhibits distinct differences from the distal half, have provided a clear structural basis to support a two-step assembly mechanism. Similar longitudinal structural variation has previously been observed in the axoneme, such as the 1–2 bridge concentrated at the proximal end (Hoops and Witman, 1983), and is presumably used to regulate the waveform of the flagella (Bui et al, 2009). Overall, there are a number of features which, taken together, suggest that basal body assembly is a tightly regulated and coordinated process. First, there is the change at the C-tubule including the structural transition at PF C1. Second, there is the attachment of luminal filaments spanning C3 to C6/7 in the C-tubule, and finally there is the relative rotation of the adjacent triplet, resulting in a change at the inter-triplet linker. In addition to key roles in assembly, these longitudinal structural variations might provide the axial polarity and spatial specificity for subsequent addition of accessory structures, such as the subdistal and distal appendages, during basal body and centriole maturation. Our structural analysis of the basal body reported here has paved the way for identification of major basal body components within the context of a 3D structure. This enables us to address fundamental questions concerning centriole and basal body biogenesis in molecular detail.
Materials and methods
Basal body purification cryo-ET data collection
Basal bodies were purified following previous published method (Snell et al, 1974; Keller et al, 2005).
The purified basal body in solution was mixed with colloidal gold (10 nm) and was applied to 300 mesh holey carbon grids (Quantifoil, Germany) and flash-frozen in liquid ethane using a Vitrobot (FEI, Inc., The Netherlands). A typical vitreous ice thickness is about 300 nm. The grids were stored in liquid nitrogen at −180°C.
ET tilt series were collected on an FEG microscope (Polara, FEI, Inc.) operating at 300 KV. The microscope was equipped with a post-column energy filter (GIF, Gatan, Inc.) and the slit width was set at 25 eV. UCSF Tomography software (Zheng et al, 2004) was used for automatic data collection. Single-axis tilt series were collected at a nominal magnification of 34 000 and images with size of 2032 × 2032 were recorded on a CCD camera (UltraCam, Gatan, Inc.) and the final pixel size of image is 6.5 Å. The specimen was tilted from −60° to +60° in 1.5° increment. The microscope defocus values were set between 9 and 24 μm. To avoid excessive radiation damage, care was taken to limit the cumulative dose on the specimen <80 e−/Å2.
3D image processing, volume average and model building
Tomographic tilt series were aligned in IMOD (Kremer et al, 1996) by using 10-nm colloid gold beads as the fiducial markers. The CTF for each tilt series was determined and corrected (Fernández et al, 2006). 3D reconstructed volumes were calculated with an iterative reconstruction algorithm (TAPIR) in Priism (Chen et al, 1996). To average sections of MT triplets from different tomogram data sets, each triplet volume is divided into a number of small segments, which are then aligned to a single common origin before averaging. We found that the longest longitudinal repeat in the triplet is 16 nm; therefore, we limited our final averaged volume to 39 nm in the longitudinal direction to contain slightly more than two repeats. Subvolumes with pixel dimension of 200 by 60 by 200 were boxed out along the basal body axis containing triplet segments about 39 nm in length and 10% overlapped with adjacent segments. Iterative subvolume alignment was carried out in Spider (Frank et al, 1996). Initially, a symmetrized triplet volume was used as a reference for alignment followed by using averaged volume from the previous round as reference for the next round of alignment. The average of the aligned subvolumes was carried out in Fourier space following an algorithm proposed by Schmid and Booth (2008). The two repeats in the averaged subvolume were further aligned and averaged to obtain the final triplet structure. A total of 1644 subvolumes from 27 tilt series were used for averaging. To assess the resolution of the final averaged structure, subvolumes were randomly split into two groups and their FSC was calculated. Since the basal body has intrinsic nine-fold symmetry where the nine triplets have different orientations, the resulting averaged triplet structure has isotropic resolution with no missing wedge artefacts. The estimated resolution is at 33 Å by using FSC=0.143 criterion (Supplementary Figure S1) (Rosenthal and Henderson, 2003). The pseudo-atomic model of the MT triplet was built based on an atomic structure of tubulin (PDB ID: 1JFF) by manually fitting into the EM density in program O (Jones et al, 1991). Care was taken to have H1-S2/H2-S3 loop and M-loop at vicinity and to avoid steric clash between laterally interacting PF. The fitting was further refined in UCSF Chimera (Pettersen et al, 2004). UCSF Chimera was used for 3D volume display and volume segmentation analysis. The contact area was estimated by assuming two density maps having surfaces within 5 Å in distance. The masses of EM densities were estimated by assuming protein density 1.41 g/cm3 (Fischer et al, 2004) after setting the density threshold by using the pseudo-atomic MT triplet model as a standard.
Supplementary Material
Acknowledgments
We thank Michael Braunfeld and Shawn Zheng for advice on tomography data collection, Lani Keller for suggestions on basal body purification, Tom Goddart for using UCSF Chimera program, Juliette Azimzadeh and members of Agard lab for discussion and comments. We also thank Richard Henderson (MRC-LMB) for critical reading of this manuscript. This work was supported by HHMI and by the Keck Laboratory for Advanced Electron Microscopy.
Author contributions: SL, WFM and DAA designed the research; SL prepared the specimen and collected the cryo-EM data; SL and JJF carried out all the image processing and map analysis; SL, JJF, WFM and DAA analysed the final result and wrote the paper.
Footnotes
The authors declare that they have no conflict of interest.
References
- Allen RD (1969) The morphogenesis of basal bodies and accessory structures of the cortex of the ciliated protozoan Tetrahymena pyriformis. J Cell Biol 40: 716–733 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amos LA (2008) The tektin family of microtubule-stabilizing proteins. Genome Biol 9: 229. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Andersen JS, Wilkinson CJ, Mayor T, Mortensen P, Nigg EA, Mann M (2003) Proteomic characterization of the human centrosome by protein correlation profiling. Nature 426: 570–574 [DOI] [PubMed] [Google Scholar]
- Anderson RG (1972) The three-dimensional structure of the basal body from the rhesus monkey oviduct. J Cell Biol 54: 246–265 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson RG, Brenner RM (1971) The formation of basal bodies (centrioles) in the Rhesus monkey oviduct. J Cell Biol 50: 10–34 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Azimzadeh J, Hergert P, Delouvée A, Euteneuer U, Formstecher E, Khodjakov A, Bornens M (2009) hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J Cell Biol 185: 101–114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bui KH, Sakakibara H, Movassagh T, Oiwa K, Ishikawa T (2009) Asymmetry of inner dynein arms and inter-doublet links in Chlamydomonas flagella. J Cell Biol 186: 437–446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carvalho-Santos Z, Machado P, Branco P, Tavares-Cadete F, Rodrigues-Martins A, Pereira-Leal JB, Bettencourt-Dias M (2010) Stepwise evolution of the centriole-assembly pathway. J Cell Sci 123(Part 9): 1414–1426 [DOI] [PubMed] [Google Scholar]
- Cavalier-Smith T (1974) Basal body and flagellar development during the vegetative cell cycle and the sexual cycle of Chlamydomonas reinhardii. J Cell Sci 16: 529–556 [DOI] [PubMed] [Google Scholar]
- Chang P, Stearns T (2000) Delta-tubulin and epsilon-tubulin: two new human centrosomal tubulins reveal new aspects of centrosome structure and function. Nat Cell Biol 2: 30–35 [DOI] [PubMed] [Google Scholar]
- Chen H, Hughes DD, Chan TA, Sedat JW, Agard DA (1996) IVE (Image Visualization Environment): a software platform for all three-dimensional microscopy applications. J Struct Biol 116: 56–60 [DOI] [PubMed] [Google Scholar]
- Chrétien D, Wade RH (1991) New data on the microtubule surface lattice. Biol Cell 71: 161–174 [DOI] [PubMed] [Google Scholar]
- Cyrklaff M, Kudryashev M, Leis A, Leonard K, Baumeister W, Menard R, Meissner M, Frischknecht F (2007) Cryoelectron tomography reveals periodic material at the inner side of subpellicular microtubules in apicomplexan parasites. J Exp Med 204: 1281–1287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deane JA, Cole DG, Seeley ES, Diener DR, Rosenbaum JL (2001) Localization of intraflagellar transport protein IFT52 identifies basal body transitional fibers as the docking site for IFT particles. Curr Biol 11: 1586–1590 [DOI] [PubMed] [Google Scholar]
- Debec A, Sullivan W, Bettencourt-Dias M (2010) Centrioles: active players or passengers during mitosis? Cell Mol Life Sci 67: 2173–2194 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dippell RV (1968) The development of basal bodies in paramecium. Proc Natl Acad Sci USA 61: 461–468 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doxsey S, Zimmerman W, Mikule K (2005) Centrosome control of the cell cycle. Trends Cell Biol 15: 303–311 [DOI] [PubMed] [Google Scholar]
- Dutcher SK (2001) The tubulin fraternity: alpha to eta. Curr Opin Cell Biol 13: 49–54 [DOI] [PubMed] [Google Scholar]
- Dutcher SK, Trabuco EC (1998) The UNI3 gene is required for assembly of basal bodies of Chlamydomonas and encodes delta-tubulin, a new member of the tubulin superfamily. Mol Biol Cell 9: 1293–1308 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fais DA, Nadezhdina ES, Chentsov YS (1986) The centriolar rim. The structure that maintains the configuration of centrioles and basal bodies in the absence of their microtubules. Exp Cell Res 164: 27–34 [DOI] [PubMed] [Google Scholar]
- Fernández JJ, Li S, Crowther RA (2006) CTF determination and correction in electron cryotomography. Ultramicroscopy 106: 587–596 [DOI] [PubMed] [Google Scholar]
- Fischer H, Polikarpov I, Craievich AF (2004) Average protein density is a molecular-weight-dependent function. Protein Sci 13: 2825–2828 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frank J, Radermacher M, Penczek P, Zhu J, Li Y, Ladjadj M, Leith A (1996) SPIDER and WEB: processing and visualization of images in 3D electron microscopy and related fields. J Struct Biol 116: 190–199 [DOI] [PubMed] [Google Scholar]
- Fromherz S, Giddings TH, Gomez-Ospina N, Dutcher SK (2004) Mutations in alpha-tubulin promote basal body maturation and flagellar assembly in the absence of delta-tubulin. J Cell Sci 117(Part 2): 303–314 [DOI] [PubMed] [Google Scholar]
- Garreau de Loubresse N, Ruiz F, Beisson J, Klotz C (2001) Role of delta-tubulin and the C-tubule in assembly of Paramecium basal bodies. BMC Cell Biol 2: 4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garvalov BK, Zuber B, Bouchet-Marquis C, Kudryashev M, Gruska M, Beck M, Leis A, Frischknecht F, Bradke F, Baumeister W, Dubochet J, Cyrklaff M (2006) Luminal particles within cellular microtubules. J Cell Biol 174: 759–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geimer S, Melkonian M (2004) The ultrastructure of the Chlamydomonas reinhardtii basal apparatus: identification of an early marker of radial asymmetry inherent in the basal body. J Cell Sci 117(Part 13): 2663–2674 [DOI] [PubMed] [Google Scholar]
- Gerdes JM, Davis EE, Katsanis N (2009) The vertebrate primary cilium in development, homeostasis, and disease. Cell 137: 32–45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giddings TH, Meehl JB, Pearson CG, Winey M (2010) Electron tomography and immuno-labeling of Tetrahymena thermophila basal bodies. Methods Cell Biol 96: 117–141 [DOI] [PubMed] [Google Scholar]
- González C, Tavosanis G, Mollinari C (1998) Centrosomes and microtubule organisation during Drosophila development. J Cell Sci 111(Part 18): 2697–2706 [DOI] [PubMed] [Google Scholar]
- Guichard P, Chrétien D, Marco S, Tassin A-M (2010) Procentriole assembly revealed by cryo-electron tomography. EMBO J 29: 1565–1572 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hinchcliffe EH, Miller FJ, Cham M, Khodjakov A, Sluder G (2001) Requirement of a centrosomal activity for cell cycle progression through G1 into S phase. Science 291: 1547–1550 [DOI] [PubMed] [Google Scholar]
- Hoops HJ, Witman GB (1983) Outer doublet heterogeneity reveals structural polarity related to beat direction in Chlamydomonas flagella. J Cell Biol 97: 902–908 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoops HJ, Wright RL, Jarvik JW, Witman GB (1984) Flagellar waveform and rotational orientation in a Chlamydomonas mutant lacking normal striated fibers. J Cell Biol 98: 818–824 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ibrahim R, Messaoudi C, Chichon FJ, Celati C, Marco S (2009) Electron tomography study of isolated human centrioles. Microsc Res Tech 72: 42–48 [DOI] [PubMed] [Google Scholar]
- Inclán YF, Nogales E (2001) Structural models for the self-assembly and microtubule interactions of gamma-, delta- and epsilon-tubulin. J Cell Sci 114(Part 2): 413–422 [DOI] [PubMed] [Google Scholar]
- Jones TA, Zou JY, Cowan SW, Kjeldgaard M (1991) Improved methods for building protein models in electron density maps and the location of errors in these models. Acta Cryst A 47(Part 2): 110–119 [DOI] [PubMed] [Google Scholar]
- Keller LC, Romijn EP, Zamora I, Yates JR, Marshall WF (2005) Proteomic analysis of isolated chlamydomonas centrioles reveals orthologs of ciliary-disease genes. Curr Biol 15: 1090–1098 [DOI] [PubMed] [Google Scholar]
- Kilburn CL, Pearson CG, Romijn EP, Meehl JB, Giddings TH, Culver BP, Yates JR, Winey M (2007) New Tetrahymena basal body protein components identify basal body domain structure. J Cell Biol 178: 905–912 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kremer JR, Mastronarde DN, McIntosh JR (1996) Computer visualization of three-dimensional image data using IMOD. J Struct Biol 116: 71–76 [DOI] [PubMed] [Google Scholar]
- Li H, DeRosier DJ, Nicholson WV, Nogales E, Downing KH (2002) Microtubule structure at 8 A resolution. Structure 10: 1317–1328 [DOI] [PubMed] [Google Scholar]
- Li JB, Gerdes JM, Haycraft CJ, Fan Y, Teslovich TM, May-Simera H, Li H, Blacque OE, Li L, Leitch CC, Lewis RA, Green JS, Parfrey PS, Leroux MR, Davidson WS, Beales PL, Guay-Woodford LM, Yoder BK, Stormo GD, Katsanis N et al. (2004) Comparative genomics identifies a flagellar and basal body proteome that includes the BBS5 human disease gene. Cell 117: 541–552 [DOI] [PubMed] [Google Scholar]
- Linck RW, Stephens RE (2007) Functional protofilament numbering of ciliary, flagellar, and centriolar microtubules. Cell Motil Cytoskeleton 64: 489–495 [DOI] [PubMed] [Google Scholar]
- Mahowald AP, Strassheim JM (1970) Intercellular migration of centrioles in the germarium of Drosophila melanogaster. An electron microscopic study. J Cell Biol 45: 306–320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mikule K, Delaval B, Kaldis P, Jurcyzk A, Hergert P, Doxsey S (2007) Loss of centrosome integrity induces p38-p53-p21-dependent G1-S arrest. Nat Cell Biol 9: 160–170 [DOI] [PubMed] [Google Scholar]
- Movassagh T, Bui KH, Sakakibara H, Oiwa K, Ishikawa T (2010) Nucleotide-induced global conformational changes of flagellar dynein arms revealed by in situ analysis. Nat Struct Mol Biol 17: 761–767 [DOI] [PubMed] [Google Scholar]
- Nicastro D, Schwartz C, Pierson J, Gaudette R, Porter ME, McIntosh JR (2006) The molecular architecture of axonemes revealed by cryoelectron tomography. Science 313: 944–948 [DOI] [PubMed] [Google Scholar]
- Nigg EA, Raff JW (2009) Centrioles, centrosomes, and cilia in health and disease. Cell 139: 663–678 [DOI] [PubMed] [Google Scholar]
- Nogales E, Whittaker M, Milligan RA, Downing KH (1999) High-resolution model of the microtubule. Cell 96: 79–88 [DOI] [PubMed] [Google Scholar]
- O’Toole ET, Giddings TH, McIntosh JR, Dutcher SK (2003) Three-dimensional organization of basal bodies from wild-type and delta-tubulin deletion strains of Chlamydomonas reinhardtii. Mol Biol Cell 14: 2999–3012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paintrand M, Moudjou M, Delacroix H, Bornens M (1992) Centrosome organization and centriole architecture: their sensitivity to divalent cations. J Struct Biol 108: 107–128 [DOI] [PubMed] [Google Scholar]
- Pelletier L, O’Toole E, Schwager A, Hyman AA, Müller-Reichert T (2006) Centriole assembly in Caenorhabditis elegans. Nature 444: 619–623 [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE (2004) UCSF Chimera—a visualization system for exploratory research and analysis. J Comput Chem 25: 1605–1612 [DOI] [PubMed] [Google Scholar]
- Piasecki BP, LaVoie M, Tam L-W, Lefebvre PA, Silflow CD (2008) The Uni2 phosphoprotein is a cell cycle regulated component of the basal body maturation pathway in Chlamydomonas reinhardtii. Mol Biol Cell 19: 262–273 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piel M, Nordberg J, Euteneuer U, Bornens M (2001) Centrosome-dependent exit of cytokinesis in animal cells. Science 291: 1550–1553 [DOI] [PubMed] [Google Scholar]
- Ringo DL (1967) Flagellar motion and fine structure of the flagellar apparatus in Chlamydomonas. J Cell Biol 33: 543–571 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Riparbelli MG, Callaini G (2011) Male gametogenesis without centrioles. Dev Biol 349: 427–439 [DOI] [PubMed] [Google Scholar]
- Rosenthal PB, Henderson R (2003) Optimal determination of particle orientation, absolute hand, and contrast loss in single-particle electron cryomicroscopy. J Mol Biol 333: 721–745 [DOI] [PubMed] [Google Scholar]
- Schmid MF, Booth CR (2008) Methods for aligning and for averaging 3D volumes with missing data. J Struct Biol 161: 243–248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Snell WJ, Dentler WL, Haimo LT, Binder LI, Rosenbaum JL (1974) Assembly of chick brain tubulin onto isolated basal bodies of Chlamydomonas reinhardi. Science 185: 357–360 [DOI] [PubMed] [Google Scholar]
- Song YH, Mandelkow E (1995) The anatomy of flagellar microtubules: polarity, seam, junctions, and lattice. J Cell Biol 128: 81–94 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sui H, Downing KH (2006) Molecular architecture of axonemal microtubule doublets revealed by cryo-electron tomography. Nature 442: 475–478 [DOI] [PubMed] [Google Scholar]
- Sui H, Downing KH (2010) Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure 18: 1022–1031 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tilney LG, Bryan J, Bush DJ, Fujiwara K, Mooseker MS, Murphy DB, Snyder DH (1973) Microtubules: evidence for 13 protofilaments. J Cell Biol 59(2 Part 1): 267–275 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vorobjev IA, Chentsov Y (1982) Centrioles in the cell cycle. I. Epithelial cells. J Cell Biol 93: 938–949 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Tsai J-W, Imai JH, Lian W-N, Vallee RB, Shi S-H (2009) Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature 461: 947–955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamashita YM, Mahowald AP, Perlin JR, Fuller MT (2007) Asymmetric inheritance of mother versus daughter centrosome in stem cell division. Science 315: 518–521 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zheng QS, Braunfeld MB, Sedat JW, Agard DA (2004) An improved strategy for automated electron microscopic tomography. J Struct Biol 147: 91–101 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.