Fig. 2. An unstable acceptor stem is isomerized to allow for CCACCA addition.
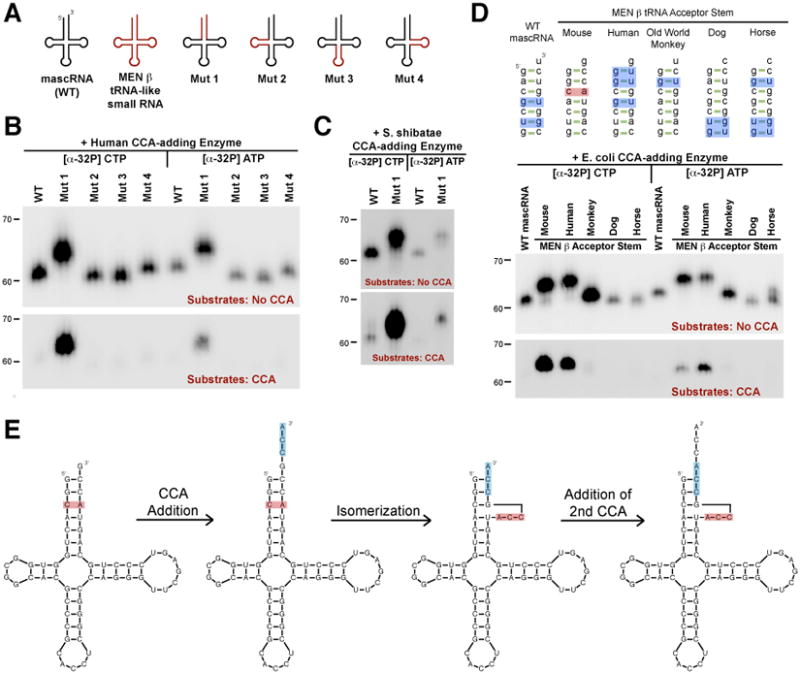
(A) Chimeric transcripts generated. (B and C) Wild-type mascRNA (denoted WT) and the chimeras were incubated in vitro with human (B) or S. shibatae (C) CCA-adding enzyme. [α-32P] CTP or [α-32P] ATP was added to assay for incorporation of each nucleotide into the unlabeled substrates. In the upper panels, substrates lacked CCA so all substrates should undergo, at minimum, one round of CCA addition, with a second round of CCA addition (yielding CCACCA) generating a 3 nucleotide shift in mobility. In the lower panels, CCA was already present so only RNAs that are CCACCA targets yield visible bands. (D) WT mascRNA or chimeras in which the acceptor stem was swapped with that of a MEN β homolog were used as substrates. Mismatches and G-U wobble base pairs are in red and blue, respectively. (E) Model for how CCACCA is added to the mouse MEN β tRNA-like small RNA.
