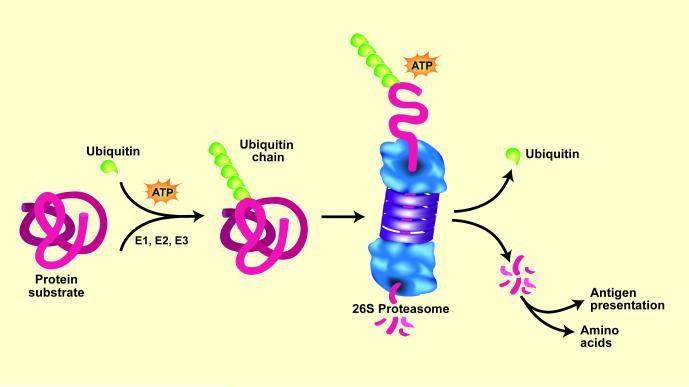
Figure 1.
The ubiquitin-proteasome system (UPS) of protein degradation. Proteins destined for degradation are conjugated to Ub by an ATP-dependent process that involves three enzymes: E1, E2, and E3. Selectivity of the protein substrates for degradation principally depends on recognition of the protein by specific E3 ligases. When a chain of five Ub proteins is attached to the protein substrate, the complex can be recognized by the 26S proteasome. This leads to release of the Ub chain, which is cleaved to individual Ubs, and unfolding of the protein substrate for injection into the proteasome and digestion to peptides. Peptidases in the cytoplasm degrade peptides into amino acids. In catabolic states, a similar process degrades muscle proteins. (Ub = ubiquitin). From Lecker, SH, Goldberg, AL and Mitch, WE; reproduced with permission from Journal of the American Society of Nephrology volume 17, pages 1807-1819, 2006.