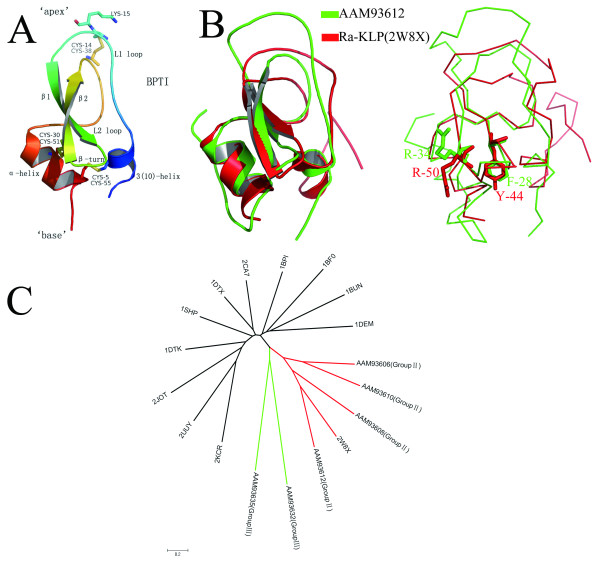
Figure 5.
Structure of classical Kunitz/BPTI protein and structural alignment of group II protein and Ra-KLP. (A), Structure of classical Kunitz/BPTI domain (PDB code 1BPI). Secondary structural elements were colored from blue (N-terminus) to red (C-terminus). The three conserved disulphide bridges and the P1 residue (Lys15 in BPTI) are shown as an atom-colored stick model. (B), Structural alignment of proteins AAM93612 (group II) and Ra-KLP (PDBID: 2W8X). The two proteins share strong similarities in the regions of 310 helix and β-turn at the "base" of the structure. In contrast, there is significant difference at the "apex" of the structure between the two proteins, due to different type indels. The superimposed structures are shown as cartoon (left) and ribbon (right). The key residues associated with channel-modulating activity are represented as stick (AAM93612: R-34, F-28; Ra-KLP: R-50, Y-44). (C), Structure-based neighbor-joining (NJ) tree of Kunitz/BPTI proteins. Six protein modes (AAM93606, AAM93608, AAM93610 and AAM93612 from group II; AAM93632 and AAM93635 from group III) were aligned with other Kunitz/BPTI protein structures (1BF0, 1BPI, 1BUN, 1DEM, 1DTK, 1DTX, 1SHP, 2CA7, 2JOT, 2KCR, 2UUY, 2W8X) using the MISTRAL online server. The root-mean-square deviation (RMSD) of pairwise structures was used to reconstruct neighbor-joining (NJ) tree. The length of each branch is proportional to the RMSD. Bar denotes 20% RMSD.