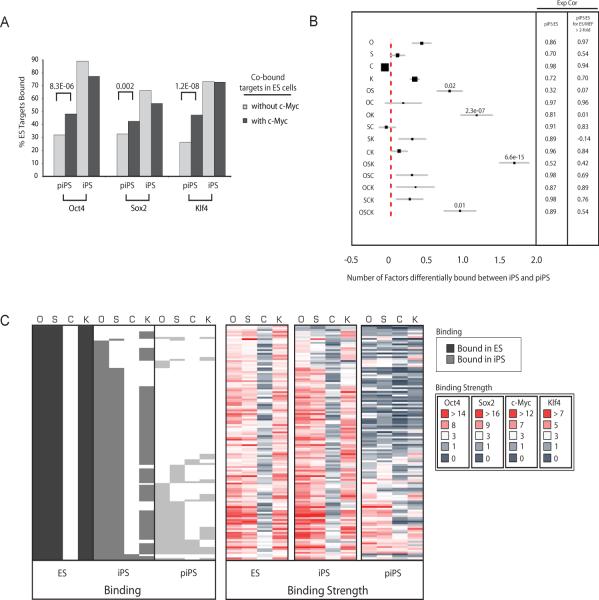
Figure 3. OSK targets are most differentially bound between iPS cells and partially reprogrammed cells.
(A) Genes bound by two, three or four factors in ES cells were divided into those co-occupied by c-Myc and those lacking c-Myc. For both groups of genes, the bar plot presents the fraction of Oct4, Sox2 or Klf4 ES targets that are bound by the respective factor in partially reprogrammed cells (piPS) or iPS cells. Significant differences for binding in piPS, computed using the difference of proportions test, are indicated.
(B) The graph displays the average number of factors differentially bound between iPS and partially reprogrammed cells for target genes grouped according to their factor association in ES cells (given to the left), within a 95% confidence interval. Filled squares reflect the relative number of genes in each target group. Significance was determined using the binomial distribution. Pearson correlation coefficients (Exp Cor) for expression between ES and partially reprogrammed cells (piPS) were computed for genes in each group and for those genes in a given group that are expressed at least two-fold higher in ES cells compared to MEFs.
(C) Binding calls and binding strength heatmaps of Oct4, Sox2, c-Myc and Klf4 in ES cells, iPS cells and partially reprogrammed cells (piPS, lightest grey) for OSK ES targets as described in Fig 1D.