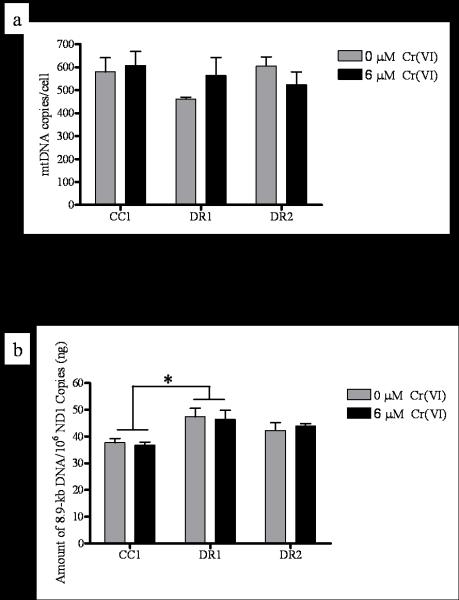
Fig. 6.
DR cells display no change in mtDNA copy number, but intrinsically less mtDNA damage. CC1, DR1, and DR2 cells were treated with 0 or 6 μM Cr(VI) for 24 h. Total DNA was extracted and quantified, (a) mtDNA and nuclear (nu) DNA copy number were determined by RT-PCR. Primers were generated for the 108-bp mitochondrial-encoded ND1 gene and an 89-bp β2 microglobulin (β2m) gene to assess mtDNA and nuDNA, respectively. Values are averages of three determinations obtained in one RT-PCR run. The copy number/cell was calculated by dividing nuDNA copy number by two (assuming the cells were diploid). mtDNA copy number was expressed as mtDNA copies/cell. (b) mtDNA damage was measured by the amplification of an 8.9-kb mtDNA target by QPCR. mtDNA damage was inversely proportional to amount of amplification , and was expressed as amount of 8.9-kb DNA/106 ND1 copies amplified. * indicates a statistically significant difference from CC1 cells, regardless of Cr(VI) treatment. Data are mean +/− SE of three experiments (0 μM Cr(VI), p = 0.0516; 6 μM Cr(VI) p=0.0430).