Abstract
The current standard for monitoring sleep in rats requires labor intensive surgical procedures and the implantation of chronic electrodes which have the potential to impact behavior and sleep. With the goal of developing a non-invasive method to determine sleep and wakefulness, we constructed a non-contact monitoring system to measure movement and respiratory activity using signals acquired with pulse Doppler radar and from digitized video analysis. A set of 23 frequency and time-domain features were derived from these signals and were calculated in 10 s epochs. Based on these features, a classification method for automated scoring of wakefulness, non-rapid eye movement sleep (NREM) and REM in rats was developed using a support vector machine (SVM). We then assessed the utility of the automated scoring system in discriminating wakefulness and sleep by comparing the results to standard scoring of wakefulness and sleep based on concurrently recorded EEG and EMG. Agreement between SVM automated scoring based on selected features and visual scores based on EEG and EMG were approximately 91% for wakefulness, 84% for NREM and 70% for REM. The results indicate that automated scoring based on non-invasively acquired movement and respiratory activity will be useful for studies requiring discrimination of wakefulness and sleep. However, additional information or signals will be needed to improve discrimination of NREM and REM episodes within sleep.
Keywords: Automated sleep scoring, Non-invasive, Movement and respiratory activity, Doppler radar, Support vector machine
1. Introduction
Accurate assessment and analysis of sleep stages is a fundamental requirement in sleep research. Rodents are often used as models in the sleep field due to their ready availability and the similarities of their sleep to human sleep (Bergmann et al., 1987). Three basic states of arousal and sleep are typically distinguished in basic sleep research: wakefulness, non-rapid eye movement sleep (NREM) and rapid eye movement sleep (REM). Determining these three arousal states in rats and other animals typically relies on recordings of the electroencephalogram (EEG) and electromyogram (EMG) and assessments of state-related changes using well-established scoring conventions.
While conventional scoring techniques yield accurate results in discriminating arousal and sleep states through the examination of electrophysiological signals obtained from animals, they also have a number of inherent limitations. These include the need for labor-intensive surgery to implant electrodes and the need to provide extensive post-surgical care to recovering animals. There is also the possibility that the recording technique (e.g., cable recording) can affect the parameter being measured and/or may limit the animal’s behavior (Tang and Sanford, 2002). Scoring the resultant EEG and EMG recordings to determine wakefulness and sleep states can also be time-consuming. These limitations indicate the need for non-invasive techniques that can record physiological parameters and that are amenable to rapid assessment of behavioral states.
A number of non-invasive approaches for assessment of behavioral state have been attempted. These include measurements of activity resulting from infrared beam breaks, frame-by-frame analysis of digital video (Pack et al., 2007) and from a pressure sensor located on the cage floor (Donohue et al., 2008; Flores et al., 2007). Each of these methods reportedly provides reasonable accuracy in distinguishing sleep from wakefulness. However, work in both humans and animals indicate that analysis of movements is not sufficient for discriminating sleep and wakefulness in all situations. In humans, actigraphy can fail during periods of low activity in wakefulness (Karlen et al., 2008) and its accuracy may decline as sleep efficiency decreases (Ancoli-Israel et al., 2003; Morgenthaler et al., 2007a; Morgenthaler et al., 2007b). In animals, our lab has shown that movement may be less discriminating of sleep for less active strains of mice (Tang et al., 2002). In addition, while detection of wakefulness and sleep is adequate for many purposes, methods need to be developed that will enable NREM and REM to be distinguished to provide broader utility and improved data across situations.
In humans, respiratory rates show state-related differences with slower, steady rates in NREM, whereas one of the hallmark signs of REM is irregular respiratory activity, in particular, during phasic REM (Pack et al., 1988). Heart rate also slows from relaxed wakefulness to NREM. It is also low during tonic REM, but there can be wide swings in heart rate during phasic REM (Pack et al., 1988). Changes in respiratory and heart rates, by themselves, may not be sufficient to clearly distinguish sleep and wakefulness. However, in combination with a measure of movement, the validity and reliability of state determination based on these parameters may be improved considerably (Karlen et al., 2008).
Doppler radar has been used to record movement in rodents (Kjellstrand et al., 1985; Marsden and King, 1979; Rose et al., 1985) and has the potential for monitoring physiologic signals including respiratory rate (Gordon and Ali, 1984; Lin, 1992; Lin, 1975) and heart rate (Lin, 1992). Doppler radar has been applied to human respiratory and heart monitoring (Staderini, 2002) and Doppler measurements of movement and respiratory activity have been used in humans to determine sleep-wake states (de Chazal et al., 2008).
In this paper, we describe a method using pulse Doppler radar for non-invasive assessment of sleep and wakefulness in rats. We utilized a 5800 MHz pulse Doppler radar sensor to non-invasively detect movement and respiratory activity, and then based on these signals, we developed an automated computer program to classify wakefulness, NREM and REM method using support vector machines (SVMs). Parallel measures of activity using digital video analysis were also obtained. The accuracy of the of non-invasively detected changes in state was determined by comparing results to those obtained by concurrent recording and scoring of sleep states based on EEG and EMG parameters. Our goal was to devise a non-invasive sleep and arousal monitoring system suitable for high-throughput screening and for assessing sleep in experimental situations (e.g., stress paradigms) that may be susceptible to confounds produced by cabling or other recording devices. The results suggest that this approach can provide a useful complementary research method for sleep research.
2. Methods
2.1. Subjects
The subjects were 7 male Wistar rats of approximately 10 weeks of age at the time of surgery. The rats were individually housed in polycarbonate cages and given ad libtitum access to food and water. The colony rooms were kept on a 12/12 light/dark cycle with lights on 07:00 to 19:00, EST. Ambient room temperature was maintained at 24.5±0.5 °C.
2.2 Surgery
The rats were implanted with two screw electrodes in the skull for recording the electroencephalogram (EEG). An additional screw electrode was placed in the skull for use as a ground. Two stainless steel wire electrodes were sutured to the dorsal neck musculature for recording the electromyogram (EMG). Two additional steel wires were implanted in the diaphragm (0.5–1 cm apart) for recording diaphragm EMG (DiaEMG) activity as a measure of respiration and to record the electrocardiogram (EKG). Leads from the recording electrodes were routed to a 9-pin miniature plug that was affixed to the skull with dental acrylic and stainless steel anchor screws.
For surgery, the rats were anesthetized with isoflurane (5% induction; 2% maintenance). Ibuprofen (15 mg/kg weight) for the relief of postoperative pain was provided in their water supply for two to three days prior to surgery and for three or more days post-surgery. The rats were allowed a minimum of 14 days to recover from surgery prior to beginning the experiment. All procedures were conducted in accordance with the National Institutes of Health Guide for the Care and Use of Experimental Animals and were approved by Eastern Virginia Medical School’s Animal Care and Use Committee (Protocols #07-013 and # 09-019).
2.3 Instruments and data collection
For recording, rats in their home cages were placed in a chamber outfitted for electrophysiological recording and a lightweight, shielded cable was attached to the miniature plug on the rat’s head. The cable was connected through a swivel commutator (Model SLC12; Plastics One, Inc. Roanoke, VA), which permitted relatively free movement of the animal. The EEG, EMG and DiaEMG signals were routed to a Grass (West Warwick, RI) Model 12 polygraph equipped with model 12A5 amplifiers. High-pass filters were set at 1 Hz and low-pass filters were set at 100 Hz for the EEG and DiaEMG and at 300Hz for the EMG. Approximately 8 h recordings were obtained from each rat beginning two h after lights on.
The biomotion sensor used was a 5800 MHz 4-channel pulse Doppler “bubble” radar unit (Model BBL; McEwan Technologies, Las Vegas, NV) with a patented modulation system that limits both the maximum and minimum detection range for distances from subjects. The output 4 channels are composed of 2 pairs of in-phase (I) and quadrature (Q) signals designed to eliminate range dependent sensing nulls. Analog filters with bandwidth of 1–22 Hz and 0.5–6 Hz were applied to 2 pairs of I/Q channels for the detection of heart beats and respiratory movement, respectively. The biomotion sensor was placed 70 cm above rat cage with its antenna pointing perpendicularly toward the cage.
Electrophysiological and biomotion signals were routed to a National Instruments 16-bit data acquisition board (NI PCI-6221) and processed at a sampling rate of 250 Hz per channel using a custom software program developed in MATLAB 7.0 (The Mathworks, Natick, MA). This program concurrently recorded both invasively derived physiological signals (EEG, EMG, DiaEMG, EKG) and non-invasively obtained signals from the biomotion sensor for subsequent comparisons of sleep-stage scoring accuracy.
Video recording was accomplished with an infrared non-contact thermal imager (Flir Systems, ThermoVision A320). This camera was used in an attempt to measure body surface temperature, but was unable to reliably detect temperature changes associated with changes in arousal state. Thus, we were limited to analysis of movement in the video.
2.4 Visual scoring
Wakefulness, NREM and REM were visually scored in 10 s epochs by two trained observers using standard EEG and EMG criteria. Wakefulness was scored based on the presence of low-voltage, fast EEG and high amplitude, tonic EMG levels, and phasic EMG bursts that could be associated with gross body movements. NREM was characterized by the presence of spindles interspersed with slow waves, lower muscle tone and no gross body movements or EEG desynchronization. REM was scored continuously during the presence of low voltage, fast EEG, theta rhythm and muscle atonia with the onset of REM occurring immediately following the last sleep spindle. All scoring was performed using SleepSign program (KISSEI COMTEC CO., LTD. Nagano 390–1293, Japan).
2.5 Doppler Radar signal processing
The I and Q channels of the quadrature radar sensor are phased 90° apart to insure that at least one of outputs is not in a null point. To avoid a phase demodulation null point and to accurately recover the bio-motion signals, output from I and Q channels must be combined. We used principal component analysis (PCA) to combine channels. PCA is a statistical method for dimensionality reduction which is commonly used to replace attributes with smaller numbers of new attributes called principal components. It computes linear combinations of the original attributes to maximally explain the variance. In this case, we reduced two dimensional data (from I and Q channels) into one. A covariance matrix between I and Q channels was calculated, and I and Q data were then projected onto an eigenvector of the covariance matrix with largest eigenvalue. Thus, the principal components resulting from the combination of I and Q channels accounted for as much of the variability within I and Q data as possible. A band pass (0.1– 12 Hz) finite impulse response (FIR) filter (Hamming windowed impulse response) with order 1024 was then applied to the PCA processed data.
2.6 Critical properties and feature extraction
The primary question for this project was whether wakefulness, NREM and REM states could be distinguished based on movement and respiratory patterns detected by Doppler radar. In the radar recordings, active wakefulness was characterized by high amplitude signals with random transient shifts. There also was increased power at low frequencies generated by large movements as a rat moved within its cage. During sleep, respiration was the major detectable movement and both NREM and REM sleep exhibited a quasi-periodic breathing pattern. Respiratory rates show slower, steady rates in NREM whereas respiration is irregular in REM, in particular, during phasic REM (Pack et al., 1988). Respiratory signals can vary in amplitude and scale with variations in sleep position and subject size. To minimize the potential effects of animal size and position, the features were extracted from normalized raw signals for each animal and were linearly scaled over all testing epochs from −1.0 to 1.0 prior to analysis.
A set of 23 frequency and time-domain features were derived from the radar sensor signal and camera (Table 1). These were calculated for successive 10 s epochs using software developed in Matlab. Features for spectral density estimates of frequency were generated using the Thompson multitaper method with a time-bandwidth product of 4 (Thomson, 1982).
Table 1.
Labels and description of the 23 features used for SVMs and automated scoring of wakefulness and sleep.
| Label | Description |
|---|---|
| SID | Standard deviation of consecutive image difference |
|
| |
| SNR _(L/H) | Respiratory signal to noise ratio |
| K_(L/H) | Kurtosis of raw signal |
| RFQ_(L/H) | Respiratory frequency |
| SAC_(L/H) | Standard deviation for autocorrelation of signal |
| KAC_(L/H) | Kurtosis for autocorrelation of signal |
| MAC_(L/H) | Mean autocorrelation of signal |
| K_PSD | Kurtosis of power spectral density |
| S_PSD | Skewness of power spectral density |
| MPL1 | Mean power spectral density (0.1–1HZ) |
| MP1-3 | Mean power spectral density (1–3HZ) |
| MP3-5 | Mean power spectral density (3–5HZ) |
| MP5-8 | Mean power spectral density (3–5HZ) |
| MP8-15 | Mean power spectral density (8–15HZ) |
| TPSD | Total Power spectral density |
| DP2AR_(L/H) | Distance from power spectral density peak in the 1–3 Hz range to the average respiratory peak position (2.1 Hz) |
(L/H): denotes whether the raw signal was from the lower analog band pass filter channel (L) or from the higher filter channels (H). Thus, labels with the L/H designation indicate two separate features. Power spectral densities (MPL1, MP1-3, MP5-8, MP8-15, TPSD), kurtosis for power spectral density (K_PSD), and skewness of power spectral density (S_PSD) were derived only from the L channel. SID was obtained from the infrared camera. All other features were obtained from the radar sensor.
Respiration during quiet wakefulness and sleep produces signals with consistent amplitudes and periodic waveforms. By comparison, movement of an animal during wakefulness increases the power of the signal at low frequencies and produces irregular waveforms. The autocorrelation (AC) function, and its frequency domain equivalent, the power spectrum, significantly reduce stationary noise, and thus, can increase efficiency in detecting periodic signals (Donohue et al., 2008). Hence, the AC was computed to characterize the periodic radar signal on each 10 sec epoch and was used for subsequent feature extraction. The signal to noise ratio (SNR), peak frequency within 1.0 – 2.1 Hz (respiratory frequency for rat, RFQ), mean power spectral density (MP) in various bands and total power spectral density (TPSD) were computed as critical features followed by obtaining additional features of the spectral shape (mean, standard deviation, skewness and kurtosis).
Large movements of the rats were also detected by video analysis. Differences in sequential images were computed by subtracting pixel to pixel changes in consecutive images. The resultant difference image was then calculated by its standard deviation among pixels.
Discriminatory power for each of the 23 features was determined using F-scores calculated using procedures outlined in (Chen and Lin, 2003). Larger F-scores indicate better discrimination.
2.7 Support vector machine
SVM is a classification tool that uses statistical learning theory (Vapnik, 2002) to maximize predictive accuracy while minimizing the problem of over-fitting to the data. Conventional neural networks based on Empirical Risk Minimization principle (ERM), which minimizes error in training data, can generalize poorly. By comparison, the SVM usually performs better in generalization due to its formulation of Structural Risk Minimization (SRM) principle which minimizes an upper bound on the expected risk (Burges, 1998). The main objective of a binary SVM is to separate data with an optimized hyperplane which maximizes the margin between two classes. SVMs are being used in a variety of biomedical and research applications including sleep-stage scoring based on EEG and EMG (Crisler et al., 2008), protein function classification (Cai et al., 2003), and the design of a brain computer interface (BCI) system (Lal et al., 2004). In this study, a soft-margin SVM was designed to automate the sleep scoring process on the features extracted from Doppler radar signals as described in Section 2.6.
2.8 Implementation and assessment of classifier
The SVM was implemented using the LIBSVM package in Matlab (Chang and Lin, 2001). A sequential minimal optimization (Platt, 1999; Zanni et al., 2006) method was used to break the optimization problem down into sub-problems thereby eliminating the need for numerical optimization at each step. All features described in Section 2.6 were scaled to within the range of −1 to 1, and were used in the SVM to automatically classify arousal states. Three binary SVM classifiers (wakefulness vs. not-wakefulness, REM vs. not-REM, NREM vs. not-NREM) were constructed and trained with selected features using one-versus-all approach for multi-classification (Abe, 2010), the final binary classification were generated according to soft scores which represent the signed distance to the decision boundary of the SVMs. The test features were presented to all three SVMs and the decision regarding state was made according to the maximum soft scores among the three classifiers.
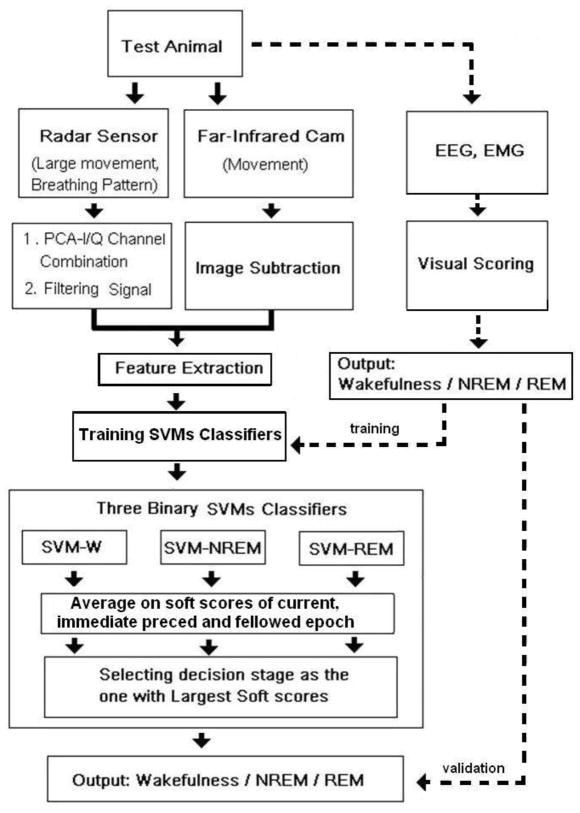
To assess the overall performance of the SVM classifier, we used a K-fold (k=10) cross-validation approach (McLachlan et al., 2004). Accordingly, a dataset consisting of roughly 56 hours of recording in 7 rats was randomly partitioned into 10 approximately equal and balanced sub-samples. In each run, one sub-sample was used as the training set and the remaining samples were used as test sets across the 7 individual rats. Visually scored sleep stages for each sample were used as the validation data for training and testing. The results of the 10 repetitions were then averaged for each rat. Based on visual scoring, the total data set contained 38.4% wakefulness, 50% NREM and 11.6% REM. A diagram of the classification procedure is shown in Figure 1.
Figure 1.
Diagram illustrating the process by which non-invasively obtained data were acquired using pulse Doppler radar and video, automatically scored and compared to visually scored records of wakefulness and sleep based on EEG and EMG.
To assess the performance of the SVM classifiers for automated determination of sleep and wakefulness, overall accuracy, positive predictive value (PPV), sensitivity (Se), and Cohen’s kappa index (CKI) were utilized.
Overall accuracy was calculated as: A=100(1-Nr/Np) where A is the percentage of overall classification accuracy, Nr denotes number of misclassified epochs, and Np denotes total number of testing epochs.
The PPV and Se were calculated by: PPV = TP/(TP+FP) and Se=TP/(TP+FN), respectively. CKI was computed as: 2*(TP*TN-FP*FN)/((TP+FN)(FN+TN)+(FP+TN)(TP+FP)). TP indicates true positives, FP indicates false positives, TN indicates true negatives, and FN indicates false negatives. N refers to the total number of 10s epochs.
The PPV measures how exclusively it classifies a certain class, e.g., a PPV of 100% means that all predicted positives are true positives. Se is a measure of the percentage of epochs which are correctly identified by the binary classifier, a Se of 100% means that the classifier recognizes all true positives. CKI (Cohen, 1960) is an index of inter-rater reliability that is commonly used to measure the level of agreement between two sets of ratings or scores. CKI values greater than 0.80 represent almost perfect agreement, whereas CKI values between 0.61 and 0.80, 0.41 and 0.60, 0.21 and 0.40, 0 and 0.20 represent substantial, moderate, fair, and slight agreement, respectively (Landis and Koch, 1977).
3. Results
3.1 Detection of Activity and Respiration
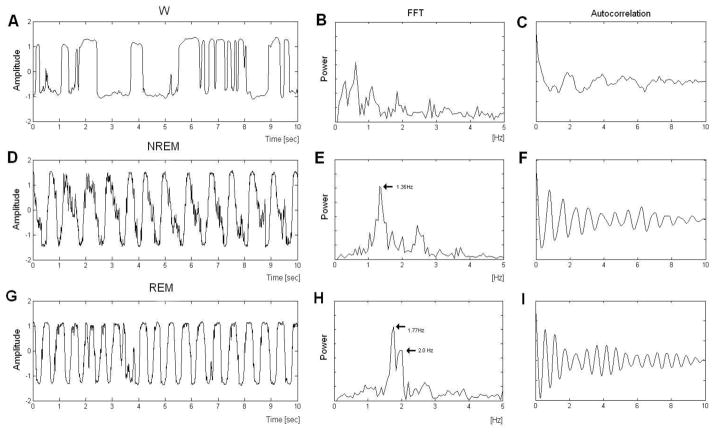
Figure 2 presents sample waveforms of signals obtained by the radar sensor in wakefulness (A), NREM (D) and REM (G). Output of the Doppler radar sensor during wakefulness was characterized by large movements and corresponding greater power in the FFT at low frequencies (Figure 2B). Output of the sensor during sleep showed cyclic oscillations that corresponded to rat chest wall motion due to respiration during NREM (Figure 2D) and REM (Figure 2G). The waveform of the respiratory signal during NREM was periodic (Figure 2D) whereas the signal during REM was irregular (Figure 2G). Mean respiratory rate during REM (2.06±0.26 Hz) was significantly higher (p<0.001) than that observed during NREM (1.59±0.1 Hz). The FFT of the respiratory signal in NREM found a prominent peak at 1.35 (Fig 3E). By comparison, the FFT of the respiratory signal in REM revealed peaks at 1.77 Hz and at 2.0 Hz (Figure 2H).
Figure 2.
Example signals obtained from the radar sensor for A. wakefulness (W) D. non-rapid eye movement sleep (NREM), and G. rapid eye movement sleep (REM). Corresponding FFTs (B, E, H) and autocorrelations (C, F, I) are shown for W, NREM, and REM, respectively.
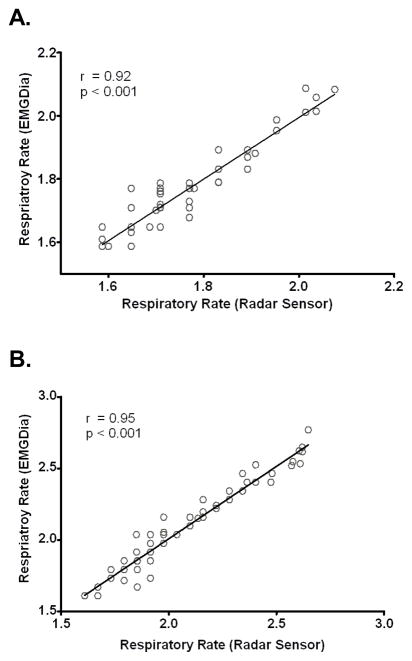
Figure 3.
Scatterplots demonstrating correlations between respiratory rates derived from the Doppler radar sensor and those obtained from the diaphragm EMG (DiaEMG). A. non-rapid eye movement sleep (NREM). B. rapid eye movement sleep (REM).
Respiratory rates derived from the Doppler radar sensor were significantly correlated with concurrently determined respiratory rates obtained from the DiaEMG using FFT peak finding methods for both NREM (Figure 3A) and REM (Figure 3B).
3.2 Sleep State Discrimination
A total of 20174 10 sec epochs (approximately 56 hours of recording; 8 hours of data in 7 rats) were visually scored by two experienced scorers based on standard EEG and EMG criteria. Overall agreement between the two scorers for all three stages was 90.36%. Agreement for each state are presented in Table 2. Some of discrepancy between scorers was due to classification bias. For example, scorer 1 assigned fewer epochs as W (7295) and REM (2132) than did scorer 2 (W: 7588; REM: 2395).
Table 2.
Agreement in scoring of wakefulness (W), NREM and REM for the two individuals who visually scored the EEG and EMG records.
| Scorer 1 | Scorer 2
|
CKI | |||
|---|---|---|---|---|---|
| W | NREM | REM | |||
| W | 7531 | 466 | 340 | 8337 (90.3%) | 0.82 |
| NREM | 1011 | 11087 | 184 | 12282(90.3%) | 0.85 |
| REM | 131 | 92 | 2214 | 2437 (90.8%) | 0.83 |
| 8673 (86.8%) | 11645(95.2%) | 2738(80.9%) | 23056 | ||
Values on the diagonal indicate agreement between the individual scorers for a given state.
Values off-diagonal indicate where the disagreements occurred. CKI: Cohen’s kappa index
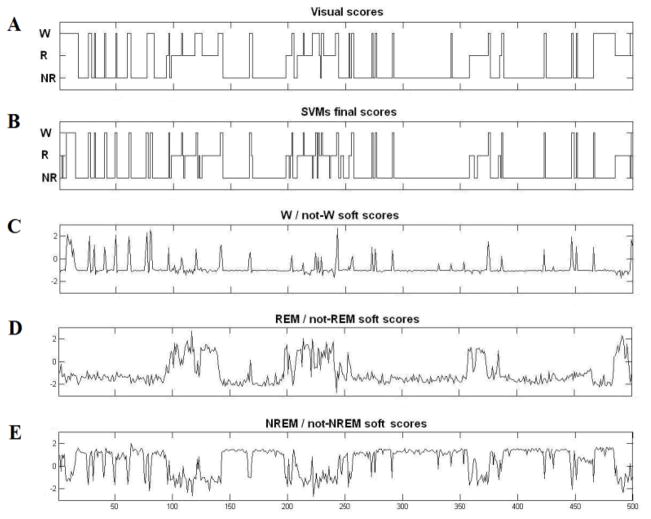
For automated scoring of wakefulness and sleep states based on the radar signal, a K-fold (k=10) cross-validation approach was utilized to evaluate the performance of SVMs in our study described as in Section 2.8. The resulting classifications were compared to the visually scored data in order to assess the performance of the automated wake-sleep stages classifier. This process is illustrated in Figure 4 which shows sleep stage classification for 500 10 sec epochs of radar sensor data made by three soft scores of the binary SVMs and the corresponding final SVM determined scores and visually determined scores. The final scores were determined by the largest soft scores among three binary SVMs.
Figure 4.
Plots of (A) visual scores and (B) SVMS final scores plotted over 500 consecutive epochs. Corresponding soft scores for the three binary SVMs ((C) wakefulness vs. not-wakefulness; (D) REM vs. not-REM; and (E) NREM vs. not-NREM) are plotted below.
Figure 5 shows the effect of adding features to the SVMs on state discrimination. We began with the feature with the greatest state discrimination based on F-score and subsequently added features with less discriminatory power until all 23 features were used. The PPV (Figure 5A) for NREM (ANOVA, F(22,138)=3.10, p<0.001) and REM (ANOVA, F(22,138)=2.06, p=0.008) increased significantly as features were added. PPV for wakefulness decreased slightly and did not significantly change as the number of features increased. By comparison, sensitivity (Figure 5B) for NREM did not vary as features were added whereas sensitivity for REM showed a steady increase as features were added (ANOVA, F(22,138)=5.41, p<0.001). Agreement increased significantly as features were added for REM (ANOVA, F(22,138)=5.72, p<0.001), NREM (ANOVA, F(22,138)=8.43, p<0.001), wakefulness (ANOVA, F(22,138)=3.22, p<0.001) and overall all-stage accuracy (ANOVA, F(22,138)=7.10, p<0.001) (Figure 5C).
Figure 5.
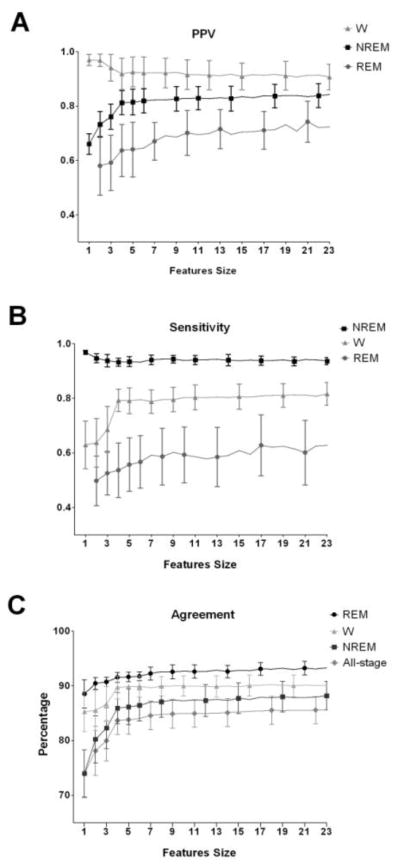
Demonstration of how adding features can alter the ability of the SVMs to classify wakefulness (W), non-rapid eye movement sleep (NREM) and rapid eye movement sleep (REM). Sequential runs of the SVM began with the feature with the greatest discrimination for a given state (based on F-scores) and subsequently added features with less discriminatory power until all 23 features were used. A. Positive predictive value (PPV), B. sensitivity and C. percentage of agreement between human rater and SVM classification are shown. W: wakefulness; NREM: non-rapid eye movement sleep; REM: rapid eye movement sleep.
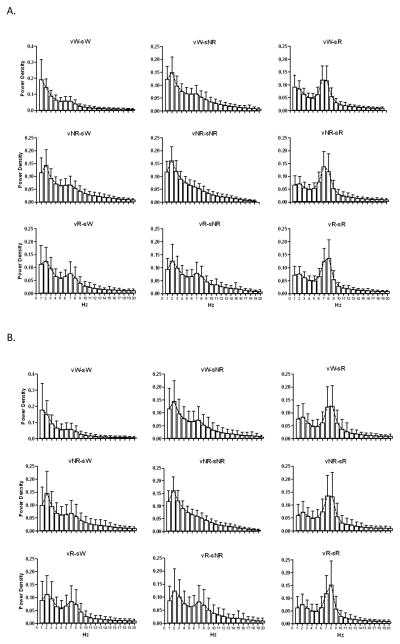
Table 3 presents indices of performance (PPV, sensitivity, CKI and accuracy) calculated for the SVMs for discriminating wakefulness, NREM and REM for all 7 rats. Agreement of the SVM automated scores to the visually scored results of the human scorers is presented in Table 4. The SVMs were trained based on Scorer 1’s results, but had similar agreement for both scorers. For each field in Table 4, we also examined the mean EEG spectral power in the 0–20 Hz range, in 1 Hz bins, across all epochs represented in individual cells (Figure 6). For instance, Figure 6A and B each have 9 panels oriented to match the cells of Table 4 showing the agreement between the SVMs and Scorer 1 and 2, respectively. One obvious observation from this figure is that visually scored epochs more closely followed the EEG spectra associated with each state than did SVM scoring which relied on variations in the respiratory signal.
Table 3.
Performance measures for the automated scoring of wakefulness, NREM and REM in each of the 7 rats.
| Rat | Wakefulness
|
NREM
|
REM
|
Acc | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| PPV | Se | CKI | PPV | Se | CKI | PPV | Se | CKI | ||
| 1 | 91.85 | 79.01 | 0.76 | 79.48 | 93.48 | 0.73 | 76.55 | 61.26 | 0.60 | 83.24 |
| 2 | 89.77 | 82.6 | 0.75 | 74.06 | 88.07 | 0.65 | 55.6 | 33.59 | 0.37 | 79.88 |
| 3 | 84.02 | 77.37 | 0.72 | 86.7 | 90.93 | 0.74 | 70.11 | 67.76 | 0.64 | 83.75 |
| 4 | 80.69 | 75.47 | 0.69 | 92.24 | 82.76 | 0.73 | 69.67 | 64.64 | 0.62 | 82.89 |
| 5 | 81.02 | 89.06 | 0.76 | 92.09 | 85.95 | 0.75 | 71.36 | 75.64 | 0.69 | 86.89 |
| 6 | 89.07 | 88.84 | 0.81 | 89.89 | 91.02 | 0.80 | 59.33 | 51.71 | 0.51 | 86.87 |
| 7 | 89.55 | 86.84 | 0.77 | 85.45 | 84.72 | 0.73 | 61.62 | 63.42 | 0.56 | 83.92 |
| Overall | 89.82 | 83.45 | 0.78 | 85.16 | 91.74 | 0.76 | 71.80 | 61.51 | 0.61 | 84.39 |
PPV: positive predictive value; Se: sensitivity (Se), CKI: Cohen’s kappa index; Acc: overall accuracy.
Table 4.
Agreement between SVM automated scores based on selected features and visual scores based on EEG and EMG.
| SVM | Scorer 1 scores
|
Scorer 2 scores
|
||||
|---|---|---|---|---|---|---|
| W(%) | NREM(%) | REM(%) | W(%) | NREM(%) | REM(%) | |
| W | 91.6±5.7 | 5.5±3.7 | 2.9±2.27 | 91.0±5.2 | 5.1±2.9 | 3.9±2.4 |
| NREM | 9.1±4.6 | 85.1±7.29 | 5.8±3.07 | 11.3±3.9 | 82.3±6.7 | 6.5±3.1 |
| REM | 15.9±8.5 | 14.9±6.76 | 69.7±13.6 | 14.8±4.1 | 12.5±4.7 | 72.7±11.4 |
Values on the diagonal indicate agreement between the SVM and individual scorers for a given state. Values off-diagonal indicate where the disagreements occurred. Values are mean ± Standard Deviation.
Figure 6.
Average EEG power spectral density for scored epochs corresponding to Table 4, v represents SVM score, s represents human scorer. vX-sY indicate epochs scored as X by SVMs and scored as Y by Scorer. A. SVMs vs Scorer 1, B. SVMs vs Scorer 2.
To further assess the accuracy of the SVMs, we also examined the agreement between automated scoring and each visual scorer on transitional epochs that were immediately preceded or immediately followed by an epoch scored as a distinct state (Table 5). Transitional epochs comprised 6.16% of the record and agreement between the SVMs and visual scoring on transitional epochs was considerably less than on non-transitional epochs.
Table 5.
Agreement between SVMs and individual visual scores for transitional epochs.
| SVM | Scorer 1 scores
|
Scorer 2 scores
|
||||
|---|---|---|---|---|---|---|
| W(%) | NREM(%) | REM(%) | W(%) | NREM(%) | REM(%) | |
| W | 7.5±4.5 | 34.8±23 | 15±11 | 6.1±3.4 | 38.8±15 | 23±7.8 |
| NREM | 24±14 | 2.9±3.53 | 4.3±4.6 | 22±12 | 3.6±3.6 | 6.1±5.5 |
| REM | 27±18 | 16±15 | 2.1±0.89 | 19±11 | 13±11 | 4.5±3.5 |
Transitional epochs are those that are immediately preceded or immediately followed by an epoch scored as a different state. Values on the diagonal indicate agreement between the SVM and individual scorers for a given transitional state. Values off-diagonal indicate where the disagreements occurred. Values are mean ± Standard Deviation.
Lastly, we examined bout length and episode numbers as measures of sleep consolidation for the SVMs and visual scoring (Table 6). For this comparison, we also calculated bout length and episode number for the SVMs determined from scores of single 10 sec epochs as well as from scoring based on the average soft score of the current and immediately preceding and following epochs. This table demonstrates that using the average soft scores produced measures of sleep consolidation that were more similar to visual scoring though REM episode duration was significantly less with both types of SVM scoring than with visual scoring.
Table 6.
Agreement between for SVMs and visual scoring for bout length and number of episodes.
| Bout length
|
Episodes number
|
Total Sleep (Min)
|
|||||||
|---|---|---|---|---|---|---|---|---|---|
| SVM | SVM (Ave) | Scorer | SVM | SVM (Ave) | Scorer | SVM | SVM (Ave) | Scorer | |
| W | 58.5±174.8 | 65.1±248.2 | 70.2.±231.1 *** | 1698 | 1274 | 1258 | 1175 | 1217 | 1295 |
| NREM | 42.2±53.0 | 99.5±98.9 | 101.4±93.5 *** | 1867 | 1322 | 1138 | 1923 | 1842 | 1692 |
| REM | 50.4±66.4 | 44.6±51.7 | 62.2±44.8 ***ΔΔΔ | 825 | 367 | 432 | 283 | 321 | 393 |
The column labeled SVM indicates bout length and episode number determined from scores of discrete 10 sec epochs. The column labeled SVM (Ave) provides bout length and episode number from scoring base on the average soft score of the current and immediately preceding and following epochs.
indicate significant difference using Kolmogorov-Smirnov test between SVM and Scorer (P<0.001),
indicate significant difference using Kolmogorov-Smirnov test between SVM (Ave) and Scorer (P<0.001). The last three columns on the right indicate how total sleep time is impacted by scoring based on SVM and SVM (Ave) compared to visual scoring.
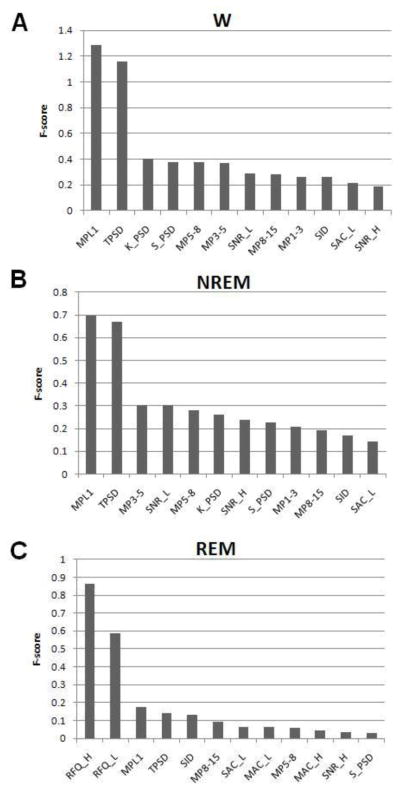
3.3 Main Discriminating Features of the SVM
Figure 7 plots the F-scores for the top 12 discriminating features used in the SVMs for wakefulness, NREM and REM. All features selected for the binary SVMs differed significantly for discriminations between wakefulness from not-wakefulness, REM from not-REM, and NREM from not-NREM (t-test, p<0.01). The features that had the largest F-scores and contributed the most to discriminating wakefulness were MPL1 and MPT. The features that contributed the most to discriminating NREM were MPL1, MPT, MP3-5, and SNR_L whereas those that contributed most to discriminating REM were RFQ and K_L, MPL1, and MP.
Figure 7.
F-scores for the top 12 features for discriminating Wakefulness (A), NREM (B), and REM (C). Description of the features are provided in Table 1.
Using only the first two top features (MPL1 and MPT), the binary SVM for discriminating wakefulness from not-wakefulness (NREM and REM) reached an 89.1% accuracy (see Figure 5C). Adding features did not noticeably improve discrimination. By comparison, using only the image data acquired from the camera (SID), yielded an accuracy of 85% discrimination of wakefulness from sleep.
The combination of three binary SVMs achieved an 81.9% agreement for all stages using only 4 features with the highest F-scores.
4. Discussion
The aim of this study was to investigate the possibility of establishing a novel, non-invasive automated sleep scoring system in rats based on gross motor activity and respiration. The results suggest that measures of activity and respiration obtained by pulse Doppler radar and analyzed using SVM procedures can provide useful estimates of the three main behavioral states of interest in sleep research.
The bio-motion radar sensor used in this study was a quadrature pulse Doppler radar which has two 90 degree phase-shifted output channels (I and Q) designed to overcome detection sensitivity to target position (Droitcour et al., 2004). In this study, PCA was used to combine signals from the I and Q channels due to superior performance over other methods (Droitcour, 2006). While the combination of the two channels improved the recovery of the bio-motion signal, periods with weak signal to noise ratio (SNR) signals were also observed, though the exact cause is unknown. Thus, despite high correlations between radar obtained signals and DiaEMG (Figure 3), some properties of the bio-motion signal, e.g., such as KAC, SAC and MAC, may not always accurately reflect the real respiratory signal with respect to breathing regularity. This was confirmed by replacing the radar derived signal with the DiaEMG signal as the source for features extraction. The result for SVMs was then improved. A more sensitive bio-motion sensor therefore could potentially provide better classification results.
Visual scoring of sleep stages based on invasively acquired physiological signals can provide reliable data regarding changes in sleep and wakefulness. Drawbacks are that visual scoring is a time-consuming process and that the results can vary depending on scorer’s training, experience and tendency. Reported agreement between scorers in published studies can range widely, e.g., from 83% to 95% (Benington et al., 1994; Louis et al., 2004; Neckelmann et al., 1994). By comparison, scorers in the current study showed an overall inter-rater agreement of 90% for all 10 sec epochs that were examined.
There has also been significant interest in the development of computer based automated scoring systems to replace visual scoring. Efforts have used a wide range of algorithms based on pattern recognition and machine learning. Most commonly used classification methods include artificial neural networks, hidden Markov models, statistic learning, and logic based methods. Reported concordance between visual and automated scoring also has a wide range, e.g., from 71% (Neckelmann et al., 1994) to 95% (Karasinski et al., 1994). However, the various methods used to determine accuracy make comparisons across studies difficult.
Relative to computer based algorithms which use invasively acquired physiological signals, our non-invasive automated system yielded high agreement with human raters in scoring for W (91.6±5.7%), NREM (85.1±7.3%), and for overall agreement for all stages (83.8± 1.87). The agreement for REM was lower (69.7±13.6%), with higher variation across the seven animals. REM accuracy may have been impacted by a number of factors including the lower number of REM epochs available in the training set and fewer distinguishing features found for REM in the radar signal.
In visual scoring, sleep and wake states are typically assigned based on characteristics that occupy the greatest percentage of a given epoch. In our automated system, some frequency and time-domain features may be applied to non stationary signals, thereby producing irregular values during mixed epochs that could present challenges to the SVM classification. Note, however, that the SVM does not rely on a strict statistical interpretation of the features, it is just looking for differentiating patterns, so it’s not a problem per se that an assumption of stationarity is violated. Indeed, the soft scoring SVM method makes a decision based on principles similar to that used in human scoring during mixed epochs and scores state based on the greatest degree of evidence during the epoch. Thus, even though mixed epochs may be more likely to be misclassified since there are some features that are based on stationary analysis, we still expect reasonable performance.
In our dataset, transitional epochs that were primarily W and were misclassified by SVMS were almost evenly distributed as REM or NREM (Table 5). By comparison, transitional epochs that were primarily NREM or REM were more likely to be misclassified by SVMs as W than as to REM from NREM or to NREM from REM (Table 5). However, given the small percentage of transitional epochs (6.16%) in our recordings, efforts made to improve the accuracy in transitional epochs will likely have a relatively smaller effect with respect to an overall improvement in performance.
Measurement of state duration and numbers of episodes are important parameters of sleep consolidation in sleep research. Based strictly on epoch to epoch evaluation, the SVMs produced more sleep episodes and shorter state durations and resulted in a record that was more fragmented than that seen with visually based scores. With visual scoring, observers may utilize information in adjacent epochs to provide context for given epoch, which may result in less fragmented records. Thus, to address this inconsistency with human scoring, we adapted the SVMs by substituting soft scores of the current epoch with average soft scores of the current and the immediately preceding and following epochs as a basis for the final score of the current epoch (Figure 1). Although this approach did not yield significant improvement in overall agreement for SVMs, there was improvement in characterizing distribution of episodes and state duration in W and NREM compared to visual scores (Table 6). However, REM duration was not significantly improved with respect to visual scoring which likely reflects the greater level of inaccuracy in detecting REM.
In future efforts, improvements to mixed epoch classification accuracy and sleep state episode consolidation could potentially be achieved by incorporating Markov model sleep-state transition likelihoods (Basner and Siebert, 2010; Kemp and Kamphuisen, 1986) into the classification decision or by identifying conditions in which there may be a mixed epoch and flagging it for subsequent manual review.
Visually scored REM epochs were consistent with the hallmark sign of greater EEG spectral power in the 5–8Hz theta wave band (see Figure 6). By comparison, automated scoring of REM epochs relied primarily on irregular respiration. Unfortunately, this parameter, as detected by the radar, was less reliable than the increase in EEG theta power during REM sleep and resulted in less accuracy for scoring REM by SVMs in comparison with visual scores. Movements produced good agreement for discriminating wakefulness; thus, the larger problem for state discrimination was related to inadequacies of the radar in discriminating sleep states when the animal was inactive.
One of the primary benefits of automated scoring is that it frees human scorers from a tedious and time consuming task. This is exemplified in the current data set; each human scorer needed approximately 8 h to visually score the 56 hours of recording. By comparison, our automated system reduced the time needed to 25–30 minutes. This consisted of 20–25 min for expert scoring to provide a training set, followed by computer processing including 290±10s for features extraction and 11.35±0.79s for SVM training and scoring. Time savings should be proportionally larger for longer recording periods.
Our initial intent was to use a Doppler radar sensor and far infrared camera to acquire heart rate, respiratory rate, movement, body surface temperature of rats. Unfortunately, a heartbeat signal was not reliably detectable using the radar sensor and potential state-related changes in surface temperature were not detectable by the infrared camera. Therefore, we were limited to using the respiratory signal and whole body movements in our efforts to develop a non-invasive automated sleep stage scoring system. However, a more sensitive radar sensor capable of detecting a heartbeat signal could further increase the accuracy of our automated scoring system. This is indicated by our finding of a 3–4% increase in the sensitivity for REM by simply by adding a heart rate feature derived from concurrently recorded EKG (data not shown).
Features extracted from time and frequency domains were chosen based on their ability to distinguish wakefulness from sleep. Large movements as a rat actively moves within its cage readily distinguish active waking and quiet periods when such activity is not present. Major features indicating large movements that we found useful included SID, MSL1, and KAC, corresponding to the sequential image difference of infrared camera, power increase in lower frequencies and decrease in the autocorrelation of radar signal, respectively.
Regularity of respiration has been used to distinguish active and quiet sleep (Sazonova et al., 2008) and was one of the primary indices used in our study. For instance, a hallmark sign of REM is irregular respiratory activity, in particular, during phasic REM (Pack et al., 1988). Autocorrelation sequences of an irregular signal have peaks at point 0 on the X axis with other signal components showing rapidly decreasing amplitudes as values on the X axis increase (see Figure 2C). By comparison, the amplitude of autocorrelation sequences for regular signals decrease more gradually (see Figure 2F). Important measures were the mean (MAC) and standard deviation (SAC) of the autocorrelation sequence, and kurtosis (K_PSD) and skewness (S_PSD) of the power spectral density which gave indices of respiratory irregularity. However, we also found an increase in respiration during REM, and the feature RFQ (respiratory frequency) provided the best discriminative power for distinguishing REM. By comparison, MAC, SAC, K_PSD and S_PSD were less discriminative between REM and other states. Since respiration in phasic REM is known to be more irregular than in tonic REM, REM epochs with our system may have been determined largely on phasic REM features.
One of the strengths of rat and mouse models is the ability to identify and utilize strains that differ on physiological and behavioral parameters. For example, Sprague-Dawley, Fischer 344, and Lewis rats differ significantly in sleep amounts (Opp and Imeri, 2001; Tang et al., 2005), activity levels (Tang et al., 2005) and Fischer and Lewis rats differ in heart rates (Baudrie et al., 2001). Breathing rates also vary among rat strains (Strohl et al., 1997). Mouse strains also differ in sleep amounts (Tang and Sanford, 2002; Veasey et al., 2000), activity levels (Tang et al., 2002), respiration (Friedman et al., 2004; Tankersley et al., 1994; Tankersley et al., 1997), and heart rates (Blizard and Welty, 1971). Being able to non-invasively measure some or all of these variables in rats and mice would provide a tremendous savings in labor. With the current Doppler radar based system, this would likely require SVMs specifically trained for each strain; however, the accuracy for comparisons across strains would need to be evaluated.
There have been a variety of attempts to non-invasively record physiological variables and use them to discriminate sleep and wake states. Recording and analyses of activity and respiration based on pressure sensing using a piezoelectric sensor has been reported to achieve 94 to 95% agreement with visual scoring of the EEG and EMG to determine wakefulness and sleep in mice (Donohue et al., 2008; Flores et al., 2007). Results with pressure sensors are in line with, or are potentially slightly better, than the 92% agreement we obtained for rats between SVM determined wake and sleep states and those determined on visually scored EEG and EMG. Periods of inactive wakefulness can be rarer in mice than in rats, which may improve sleep and wakefulness discrimination based on measures of activity. However, activity level can vary with strain and the correlation between inactivity and sleep is better for high active BALB/cJ mice than in low active DBA/2J mice (Tang and Sanford, 2002).
Pressure sensing and Doppler radar system are able to record subtle movement such as grooming and breathing movement in animals and can produce acceptable levels of discrimination of wakefulness and sleep. However, both methods have lower ability to distinguish REM from NREM, suggesting that additional signals or parameters are needed to improve the detection of REM. We were able to achieve improved distinguishing REM from NREM by adding heartbeat features. This suggests that a modification that could improve the radar’s ability to detect heartbeat provide a significant improvement in distinguishing REM from NREM as non-invasive sleep scoring system. Doppler methods have been developed to detect heart rate in humans (Lin, 1992) suggesting that applications in larger animals that utilize heart movement to improve REM detection would be possible.
The present work was based on a prototype Doppler radar sensor from a company that licenses radar technology (http://www.getradar.com/Prices2.htm) and there does not appear to currently be an off-the shelf radar product that operates similarly. Those interested in pursuing Doppler sensing in rodents could contact the company that supplied ours. Others have built their own radar circuits to perform similar sensing tasks (Fletcher and Kulkarni, 2010; Jang et al., 2008; Zito et al., 2008). In general, the technology is not expensive, so the primary costs for a system similar to the one described here would be for development plus any patent licensing fees. These costs would scale down substantially with increased volume with the result that the core technology could be quite economical.
In conclusion, our results indicate that automated scoring based on non-invasively acquired movement and respiratory activity can reliably discriminate wakefulness and sleep. However, additional information or signals will be needed to improve discrimination of NREM and REM episodes within sleep.
Highlights.
We constructed a non-contact monitoring system to measure movement and respiratory activity.
Based on these signals, we developed a method for automated scoring of wakefulness, and sleep using a support vector machine.
Agreement between automated and visual scored sleep was 91% for wakefulness, 84% for non-rapid eye movement sleep and 70% for rapid eye movement sleep.
Automated scoring based on movement and respiratory activity can discriminate wakefulness and sleep.
Acknowledgments
This work was supported by NIH research grants RR20816, MH64827 and MH61716.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Abe S. Support vector machines for pattern classification. Springer-Verlag Inc; New York: 2010. [Google Scholar]
- Ancoli-Israel S, Cole R, Alessi C, Chambers M, Moorcroft W, Pollak CP. The role of actigraphy in the study of sleep and circadian rhythms. Sleep. 2003;26:342–92. doi: 10.1093/sleep/26.3.342. [DOI] [PubMed] [Google Scholar]
- Basner M, Siebert U. Markov processes for the prediction of aircraft noise effects on sleep. Med Decis Making. 2010;30:275–89. doi: 10.1177/0272989X09342751. [DOI] [PubMed] [Google Scholar]
- Baudrie V, Laude D, Chaouloff F, Elghozi JL. Genetic influences on cardiovascular responses to an acoustic startle stimulus in rats. Clin Exp Pharmacol Physiol. 2001;28:1096–9. doi: 10.1046/j.1440-1681.2001.03593.x. [DOI] [PubMed] [Google Scholar]
- Benington JH, Kodali SK, Heller HC. Scoring transitions to REM sleep in rats based on the EEG phenomena of pre-REM sleep: an improved analysis of sleep structure. Sleep. 1994;17:28–36. doi: 10.1093/sleep/17.1.28. [DOI] [PubMed] [Google Scholar]
- Bergmann BM, Winter JB, Rosenberg RS, Rechtschaffen A. NREM sleep with low-voltage EEG in the rat. Sleep. 1987;10:1–11. doi: 10.1093/sleep/10.1.1. [DOI] [PubMed] [Google Scholar]
- Blizard DA, Welty R. Cardiac activity in the mouse: strain differences. J Comp Physiol Psychol. 1971;77:337–44. doi: 10.1037/h0031655. [DOI] [PubMed] [Google Scholar]
- Burges CJC. A tutorial on support vector machines for pattern recognition. Data mining and knowledge discovery. 1998;2:121–67. [Google Scholar]
- Cai C, Wang W, Sun L, Chen Y. Protein function classification via support vector machine approach. Mathematical biosciences. 2003;185:111–22. doi: 10.1016/s0025-5564(03)00096-8. [DOI] [PubMed] [Google Scholar]
- Chang C-C, Lin C-J. LIBSVM: a library for support vector machines. Software. 2001 available at http://www.csie.ntu.edu.tw/~cjlin/libsvm.
- Chen Y-W, Lin C-J. Combining SVMs with Various Feature Selection Strategies. 2003 URL http://www.csie.ntu.edu.tw/~cjlin/papers/features.pdf.
- Cohen J. A coefficient of agreement for nominal scales. Educational and Psychological Measurement. 1960;20:37–46. [Google Scholar]
- Crisler S, Morrissey MJ, Anch AM, Barnett DW. Sleep-stage scoring in the rat using a support vector machine. J Neurosci Methods. 2008;168:524–34. doi: 10.1016/j.jneumeth.2007.10.027. [DOI] [PubMed] [Google Scholar]
- de Chazal P, O’Hare E, Fox N, Heneghan C. Assessment of sleep/wake patterns using a non-contact biomotion sensor. Conf Proc IEEE Eng Med Biol Soc. 2008;2008:514–7. doi: 10.1109/IEMBS.2008.4649203. [DOI] [PubMed] [Google Scholar]
- Donohue KD, Medonza DC, Crane ER, O’Hara BF. Assessment of a non-invasive high-throughput classifier for behaviours associated with sleep and wake in mice. Biomed Eng Online. 2008;7:14. doi: 10.1186/1475-925X-7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Droitcour A, Boric-Lubecke O, Lubecke V, Lin J, Kovacs G. Range correlation and I/Q performance benefits in single-chip silicon Doppler radars for noncontact cardiopulmonary monitoring. Microwave Theory and Techniques, IEEE Transactions on. 2004;52:838–48. [Google Scholar]
- Droitcour AD. PhD Thesis. Stanford University; CA: 2006. Non-contact measurement of heart and respiration rates with a single-chip microwave doppler radar. [Google Scholar]
- Fletcher RR, Kulkarni S. Clip-on wireless wearable microwave sensor for ambulatory cardiac monitoring. Conf Proc IEEE Eng Med Biol Soc. 2010;2010:365–9. doi: 10.1109/IEMBS.2010.5627972. [DOI] [PubMed] [Google Scholar]
- Flores AE, Flores JE, Deshpande H, Picazo JA, Xie XS, Franken P, Heller HC, Grahn DA, O’Hara BF. Pattern recognition of sleep in rodents using piezoelectric signals generated by gross body movements. IEEE Trans Biomed Eng. 2007;54:225–33. doi: 10.1109/TBME.2006.886938. [DOI] [PubMed] [Google Scholar]
- Friedman L, Haines A, Klann K, Gallaugher L, Salibra L, Han F, Strohl KP. Ventilatory behavior during sleep among A/J and C57BL/6J mouse strains. J Appl Physiol. 2004;97:1787–95. doi: 10.1152/japplphysiol.01394.2003. [DOI] [PubMed] [Google Scholar]
- Gordon CJ, Ali JS. Measurement of ventilatory frequency in unrestrained rodents using microwave radiation. Respir Physiol. 1984;56:73–9. doi: 10.1016/0034-5687(84)90131-2. [DOI] [PubMed] [Google Scholar]
- Jang B-J, Wi S-U, Yook J-G, Lee M-Q, Lee K-J. Wireless Bio-radar Sensor for Heartbeat and Respiration Detection. Progress In Electromagnetics Research C. 2008;5:149–68. [Google Scholar]
- Karasinski P, Stinus L, Robert C, Limoge A. Real-time sleep-wake scoring in the rat using a single EEG channel. Sleep. 1994;17:113. doi: 10.1093/sleep/17.2.113. [DOI] [PubMed] [Google Scholar]
- Karlen W, Mattiussi C, Floreano D. Improving actigraph sleep/wake classification with cardio-respiratory signals. Conf Proc IEEE Eng Med Biol Soc. 2008;2008:5262–5. doi: 10.1109/IEMBS.2008.4650401. [DOI] [PubMed] [Google Scholar]
- Kemp B, Kamphuisen HA. Simulation of human hypnograms using a Markov chain model. Sleep. 1986;9:405–14. doi: 10.1093/sleep/9.3.405. [DOI] [PubMed] [Google Scholar]
- Kjellstrand P, Holmquist B, Jonsson I, Romare S, Mansson L. Effects of organic solvents on motor activity in mice. Toxicology. 1985;35:35–46. doi: 10.1016/0300-483x(85)90130-1. [DOI] [PubMed] [Google Scholar]
- Lal TN, Schroder M, Hinterberger T, Weston J, Bogdan M, Birbaumer N, Scholkopf B. Support vector channel selection in BCI. Biomedical Engineering, IEEE Transactions on. 2004;51:1003–10. doi: 10.1109/TBME.2004.827827. [DOI] [PubMed] [Google Scholar]
- Landis JR, Koch GG. The measurement of observer agreement for categorical data. Biometrics. 1977;33:159–74. [PubMed] [Google Scholar]
- Lin JC. Microwave sensing of physiological movement and volume change: a review. Bioelectromagnetics. 1992;13:557–65. doi: 10.1002/bem.2250130610. [DOI] [PubMed] [Google Scholar]
- Lin JC. Noninvasive microwave measurement of respiration. Proc IEEE. 1975;63:1530. [Google Scholar]
- Louis RP, Lee J, Stephenson R. Design and validation of a computer-based sleep-scoring algorithm. J Neurosci Methods. 2004;133:71–80. doi: 10.1016/j.jneumeth.2003.09.025. [DOI] [PubMed] [Google Scholar]
- Marsden CA, King B. The use of Doppler shift radar to monitor physiological and drug induced activity patterns in the rat. Pharmacol Biochem Behav. 1979;10:631–5. doi: 10.1016/0091-3057(79)90312-5. [DOI] [PubMed] [Google Scholar]
- McLachlan GJ, Do KA, Ambroise C. Analyzing Microarray Gene Expression Data. Wiley; 2004. [Google Scholar]
- Morgenthaler T, Alessi C, Friedman L, Owens J, Kapur V, Boehlecke B, Brown T, Chesson A, Jr, Coleman J, Lee-Chiong T, Pancer J, Swick TJ. Practice parameters for the use of actigraphy in the assessment of sleep and sleep disorders: an update for 2007. Sleep. 2007a;30:519–29. doi: 10.1093/sleep/30.4.519. [DOI] [PubMed] [Google Scholar]
- Morgenthaler TI, Lee-Chiong T, Alessi C, Friedman L, Aurora RN, Boehlecke B, Brown T, Chesson AL, Jr, Kapur V, Maganti R, Owens J, Pancer J, Swick TJ, Zak R. Practice parameters for the clinical evaluation and treatment of circadian rhythm sleep disorders. An American Academy of Sleep Medicine report. Sleep. 2007b;30:1445–59. doi: 10.1093/sleep/30.11.1445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neckelmann D, Olsen OE, Fagerland S, Ursin R. The reliability and functional validity of visual and semiautomatic sleep/wake scoring in the Moll-Wistar rat. Sleep. 1994;17:120–31. doi: 10.1093/sleep/17.2.120. [DOI] [PubMed] [Google Scholar]
- Opp MR, Imeri L. Rat strains that differ in corticotropin-releasing hormone production exhibit different sleep-wake responses to interleukin 1. Neuroendocrinology. 2001;73:272–84. doi: 10.1159/000054644. [DOI] [PubMed] [Google Scholar]
- Pack AI, Galante RJ, Maislin G, Cater J, Metaxas D, Lu S, Zhang L, Von Smith R, Kay T, Lian J, Svenson K, Peters LL. Novel method for high-throughput phenotyping of sleep in mice. Physiol Genomics. 2007;28:232–8. doi: 10.1152/physiolgenomics.00139.2006. [DOI] [PubMed] [Google Scholar]
- Pack AI, Kline LR, Hendricks JC, Morrison AR. Control of respiration during sleep. In: Fishman AP, editor. Pulmonary Diseases and Disorders. McGraw-Hill; New York: 1988. pp. 145–61. [Google Scholar]
- Platt JC. Advances in Kernel Methods-Support Vector Learning. MIT Press; Cambridge, MA: 1999. Sequetial minimal optimization: A fast algorithm for training support vector machines. [Google Scholar]
- Rose FD, Dell PA, Love S. Doppler shift radar monitoring of activity of rats in a behavioural test situation. Physiol Behav. 1985;35:85–7. doi: 10.1016/0031-9384(85)90175-1. [DOI] [PubMed] [Google Scholar]
- Sazonova NA, Sazonov EE, Tan B, Schuckers SAC. IEEE. 2008. Sleep State Scoring in Infants from Respiratory and Activity Measurements; pp. 2462–5. [DOI] [PubMed] [Google Scholar]
- Staderini EM. UWB radars in medicine. IEEE Aerospace and Electronics Systems Magazine. 2002;17:13–8. [Google Scholar]
- Strohl KP, Thomas AJ, St Jean P, Schlenker EH, Koletsky RJ, Schork NJ. Ventilation and metabolism among rat strains. J Appl Physiol. 1997;82:317–23. doi: 10.1152/jappl.1997.82.1.317. [DOI] [PubMed] [Google Scholar]
- Tang X, Liu X, Yang L, Sanford LD. Rat strain differences in sleep after acute mild stressors and short-term sleep loss. Behav Brain Res. 2005;160:60–71. doi: 10.1016/j.bbr.2004.11.015. [DOI] [PubMed] [Google Scholar]
- Tang X, Orchard SM, Sanford LD. Home cage activity and behavioral performance in inbred and hybrid mice. Behav Brain Res. 2002;136:555–69. doi: 10.1016/s0166-4328(02)00228-0. [DOI] [PubMed] [Google Scholar]
- Tang X, Sanford LD. Telemetric recording of sleep and home cage activity in mice. Sleep. 2002;25:691–9. [PubMed] [Google Scholar]
- Tankersley CG, Fitzgerald RS, Kleeberger SR. Differential control of ventilation among inbred strains of mice. Am J Physiol. 1994;267:R1371–7. doi: 10.1152/ajpregu.1994.267.5.R1371. [DOI] [PubMed] [Google Scholar]
- Tankersley CG, Fitzgerald RS, Levitt RC, Mitzner WA, Ewart SL, Kleeberger SR. Genetic control of differential baseline breathing pattern. J Appl Physiol. 1997;82:874–81. doi: 10.1152/jappl.1997.82.3.874. [DOI] [PubMed] [Google Scholar]
- Thomson DJ. Spectrum estimation and harmonic analysis. Proceedings of the IEEE. 1982;70:1055–96. [Google Scholar]
- Vapnik VN. An overview of statistical learning theory. Neural Networks, IEEE Transactions on. 2002;10:988–99. doi: 10.1109/72.788640. [DOI] [PubMed] [Google Scholar]
- Veasey SC, Valladares O, Fenik P, Kapfhamer D, Sanford L, Benington J, Bucan M. An automated system for recording and analysis of sleep in mice. Sleep. 2000;23:1025–40. [PubMed] [Google Scholar]
- Zanni L, Serafini T, Zanghirati G. Parallel Software for Training Large Scale Support Vector Machines on Multiprocessor Systems. Journal of Machine Learning Research. 2006:1467–92. [Google Scholar]
- Zito D, Pepe D, Mincica M, Zito F, De Rossi D, Lanata A, Scilingo EP, Tognetti A. Wearable system-on-a-chip UWB radar for contact-less cardiopulmonary monitoring: present status. Conference proceedings: Annual International Conference of the IEEE Engineering in Medicine and Biology Society IEEE Engineering in Medicine and Biology Society Conference. 2008;2008:5274–7. doi: 10.1109/IEMBS.2008.4650404. [DOI] [PubMed] [Google Scholar]