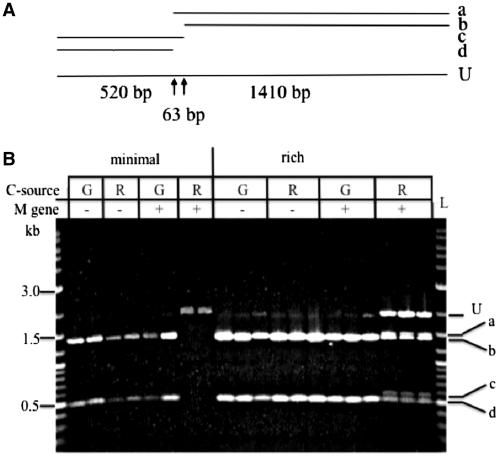
Figure 6.
Complete in vivo modification of pMC63 by rhaBp-driven fnuDIIM. (A) Methylated (protected, uncut) substrate (U) and products (a–d) expected from digestion of AclI-linearized pMC63 with BstUI. The BstUI sites (CGCG; arrows) will be protected from digestion when modified by M.FnuDII (CGCG → mCGCG). Fragments (b) and (d) are expected from a complete digest; fragments (a) and (c) may be expected in partial digests, as may occur when the plasmid is partially modified. A 63-bp central fragment is also expected from complete digestion but runs off the gel in (B). (B) pMC63 was prepared from cultures of ER3239 (M gene −) or ER3240 (M gene +). Cultures were grown in M9 (minimal, duplicates grown) or RB (rich, triplicates grown) supplemented with 0.2% sugar: glucose (G) or rhamnose (R) to repress or induce expression of fnuDIIM. The extent of in vivo modification was tested by BstUI digestion of AclI-linearized plasmid DNA prepared from the cultures and interpreted according to the diagram in panel A. Size standards on each side are the 2-log ladder (L); selected bands are labeled with size in kb. The gel system was 1% agarose with Tris–borate buffer containing ethidium bromide. Higher cell density in RB accounts for higher yields in plasmid preps. The low-intensity band at the position of ‘U’ in the rich broth cultures G−, R− and G+ (which all should be unmodified at BstUI sites) presumably are linears that result from BstUI cleavage of plasmid that escaped AclI digestion.