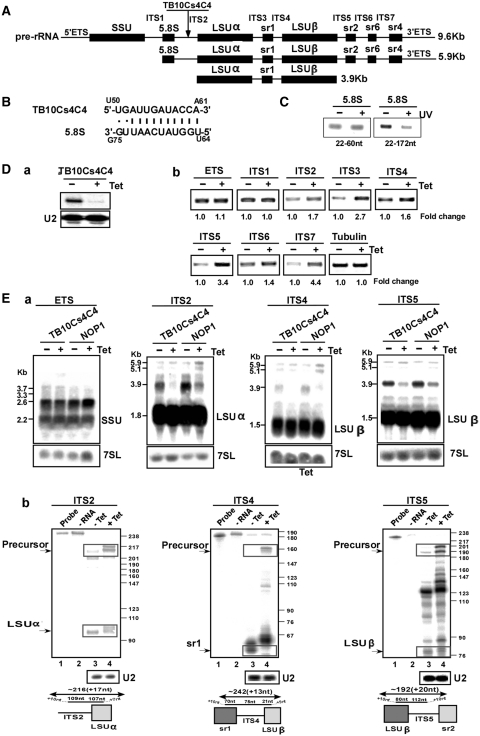
Figure 5.
The role of TB10Cs4C4 in rRNA processing. (A) Schematic representation of the pre-rRNA. The coding sequences are shown in black. The identity of the intergenic regions is given. (B) Proposed interaction domain between the snoRNA and 5.8S rRNA. (C) Mapping the interaction domain of TB10Cs4C4 with 5.8S rRNA. RT–PCR along the 5.8S RNA. cDNA was prepared from the affinity selected RNA of untreated cells or after treatment with AMT and UV cross-linking. The cDNA was subjected to PCR using the set of primers indicated in the panel, and the PCR products were separated on a 1% agarose gel and stained with ethidium bromide. (D-a) Silencing of TB10Cs4C4. Total RNA (10 µg) before and after 3 days of induction was subjected to primer extension with primer specific to the snoRNA. The level of RNA was determined using U2-specific probe. (D-b) RT–PCR to examine rRNA processing defects under TB10Cs4C4 silencing. RNA from uninduced cells or cells after 3 days of induction was used to prepare cDNA. cDNA was amplified with primers situated in the intergenic regions specified in Supplementary Figure S1. The level of tubulin mRNA was used to control the level of RNA in each sample. The identity of the intergenic regions is indicated. (E) Monitoring the accumulation of rRNA precursors in TB10Cs4C4 silenced cells. (E-a) Northern analysis. Total RNA was extracted from cells carrying the TB10Cs4C4 RNAi construct without induction (−Tet), and after 3 days of induction (+Tet) and separated on a 1.2% agarose gel containing 2.2 M formaldehyde. The RNA was blotted and hybridized with indicated probe. The 7SL RNA probe was used to control the amount of RNA of each sample. The marker is indicated in Kb. (E-b) RNase protection to detect rRNA processing defects. RNA from cells expressing the TB10Cs4C4 silencing construct was used for RNase protection assays with the different probes, as indicated. The products were separated on a 6% denaturing gel. The structure of the antisense probe is schematically presented. The precursor and the mature snoRNAs are boxed. The sizes of the molecular weight marker, pBR322 DNA-MspI digest, are indicated. In each panel: Lane 1, probe; lane 2, protection assay in the absence of RNA; lane 3, RNA from uninduced cells; lane 4, RNA from cells after 3 days of induction.