Abstract
DNA replication initiation proteins (Reps) are subjected to degradation by cellular proteases. We investigated how the formation of nucleoprotein complex, involving Rep and a protease, affects Rep degradation. All known Escherichia coli AAA+ cytosolic proteases and the replication initiation protein TrfA of the broad-host-range plasmid RK2 were used. Our results revealed that DNA influences the degradation process and that the observed effects are opposite and protease specific. In the case of ClpXP and ClpYQ proteases, DNA abolishes proteolysis, while in the case of ClpAP and Lon proteases it stimulates the process. ClpX and ClpY cannot interact with DNA-bound TrfA, while the ClpAP and Lon activities are enhanced by the formation of nucleoprotein complexes involving both the protease and TrfA. Lon has to interact with TrfA before contacting DNA, or this interaction can occur with TrfA already bound to DNA. The TrfA degradation by Lon can be carried out only on DNA. The absence of Lon results with higher stability of TrfA in the cell.
INTRODUCTION
In bacteria, many important cellular processes are mediated by AAA+ proteases (1,2). These proteolytic machines consist of two functional units acting together: an ATP-dependent unfoldase that is a molecular chaperone belonging to the AAA+ (ATPase associated with various cellular activities) protein superfamily, and a peptidase unit forming the proteolytic chamber accessible to unfolded substrate proteins (3). In Escherichia coli, four cytosolic AAA+ proteases have been identified to date: ClpXP, ClpAP, ClpYQ (also referred to as HslUV) and Lon (1). Hexameric Lon consists of unfoldase and peptidase units within a single polypeptide chain (4). For the other proteases, the ATPase unit forms a hexameric ring that interacts with the multimeric peptidase (3). The specificity of the ATPase unit is a key factor preventing uncontrolled cellular protein degradation. The recognition is based either on detection of hydrophobic stretches within the substrate protein or by binding to specific motifs called degrons and it can be affected by adaptor proteins (5–7).
DNA replication is one of the fundamental cellular processes affected by proteases and molecular chaperones. Chaperone proteins take part in the activation of replication initiation proteins or in the remodeling of nucleoprotein complexes, while proteases are involved in the degradation of replication initiators (Reps). In E. coli molecular chaperones convert Rep multimers to monomers which allows the replication initiators to specifically interact with direct repeats (iterons) located within replication origins (8–11). The molecular chaperones able to process Rep proteins are often involved in proteolysis of these proteins, functioning as unfoldase units of ATP-dependent proteases. The action of these proteases limits the half-life of Rep initiator proteins in cells which is important for replication initiation. It has been demonstrated that the initiator proteins of bacteriophages lambda and Mu and plasmid RK2 are proteolysed by E. coli ClpXP (12–14). Protease ClpAP degrades Rep of plasmid P1 (15). It was also shown that eukaryotic replication proteins are affected by chaperones and proteases (16,17).
The RK2 plasmid (for a recent review see 18) is a 60 kbp broad-host-range replicon which replicates and is stably maintained in essentially all Gram-negative bacteria tested. The binding of the RK2 replication initiation protein TrfA to the iterons located within RK2 origin of replication (oriV) leads, through subsequent steps, to the formation of the replication complex. Similar to other plasmid and eukaryotic initiators, wild-type TrfA contains two winged helix (WH) domains (13,19) and exists largely in the form of a dimer, however it strongly binds iteron DNA as a monomer (20). Dimeric wild-type TrfA is inert in the in vitro replication assays unless it is incubated with the E. coli ClpX or E. coli ClpB and DnaK, DnaJ and GrpE chaperones (8,9). This increases the proportion of TrfA monomers and, therefore, the ability of this protein to bind to iterons. TrfA dimers inhibit RK2 replication by origin pairing process termed ‘handcuffing’ (18). The proteolysis of both forms of TrfA should be important, given that this replication initiator does not have the autorepression regulatory mechanism which is common for most known Rep proteins (19). It has been demonstrated that ClpXP degrades TrfA both in vivo and in vitro, however this proteolysis is limited to the dimeric form of the protein (13).
In this work we investigated the effects of DNA on the activity of all E. coli cytosolic AAA+ proteases toward the replication initiator TrfA. Our experiments were focused on elucidation of the mechanisms accounting for these effects.
MATERIALS AND METHODS
Bacterial strains, oligonucleotides and plasmids
Escherichia coli strains used in this study were C600 and its derivatives: SG12050 (C600 clpP::Cmr) and ATC12017 (C600 lon510) (21). Oligonucleotides used for cloning of the E. coli lon gene were: 5′-AATCCTGAGCGTTCTGAACGCATT-3′ and 5′-GAGCATGCCTATTTTGCAGTCACAACC-3′. DNA fragments used in the SPR experiments were obtained by hybridization of two complementary single-stranded oligonucleotides which were purchased from a commercial source (Thermo Scientific). 5′-terminally biotinylated double-stranded DNA fragments used for the SPR analysis were: frag1 (5′-CCTGCGGTATTGACACTTGAGGGGCGCGACTACTGACAGATGA GGGGCGCGATCCTTGACACTTGAGGGGCAGAGTGATGACAGATGAGGGGCGCACCTATTGACATTTGAGGGGCTGTCCACAGGCAG-3′) and frag2 (5′-AGCTCACAATTCCACACAACAT ACGAGCCGGAAGCATAAAGTGTAAAGCCTGGGGTGCCTAATGAGTGAGCTAACTCACATTAATTGCGTTGCGCTCACTGCCCGCTTTCCAGTCGGGAAACCTGTCGT-3′). Plasmids pAT30 carrying the genes for His6-TrfA-33 or its mutant variants His6-TrfA-33 G254D/S267L and His6-TrfA-33 P151S/G254D/S267L were used for purification of TrfA. Plasmids pBAD24ClpX and pSK20 were used for purification of ClpX and ClpP, respectively. Plasmid pUHE21-2fdΔ12-ClpA for overproduction and purification of ClpA was a kind gift from Prof. Bernd Bukau (ZMBH, Heidelberg). Plasmids pET12b-His6-ClpY and pET12-ClpQ-His6 were kindly provided by Prof. Matthias Bochtler (IMCB, Warsaw). The plasmid used for purification of Lon protease (pBADLon) was constructed by inserting the PCR-amplified fragment containing lon gene between NcoI and SphI sites of pBAD24. Plasmid pBK20 is a pUC19 vector with five oriV iterons inserted into its BamHI site (22) and was purified from E. coli cells by alkaline lysis and isopropanol precipitaion followed by two centrifugations in cesium chloride gradient. Plasmids pRR10 and pTJS42 are minireplicons of the RK2 plasmid (22).
Protein purification and determination of proteolytic activity
Experiments described in this study utilized highly purified proteins (95% or higher purity). All TrfA preparations used in the experiments were N-terminally histidine-tagged 33 kDa versions of the protein. Purification of TrfA variants including wt TrfA, TrfA G254D/S267L and TrfA P151S/G254D/S267L was performed essentially as described in (20). Published protocols were applied for purification of ClpA (23), ClpP (24), ClpY (25), ClpQ (26) and Lon (27). ClpX was purified using a combination of ion-exchange chromatography methods described previously (13). The proteolytic activity of Clp and Lon proteins was measured using modified method described previously (24) with α-casein as a substrate for ClpAP, ClpYQ and Lon, and λO protein for ClpXP. Proteolysis of excess substrate was carried out for 15 min as described for the in vitro proteolysis assay. Specific activities for both components of the Clp proteases were measured by keeping one component limiting and using a saturating amount of the other component. The amount of degraded substrate was estimated after SDS–PAGE, Coomasie staining and densitometry analysis. A unit of activity was defined as the degradation of 1 mg of substrate protein per hour.
In vitro proteolysis assay
Standard proteolysis reaction had a volume of 25 or 50 µl and contained 1.5 µg TrfA and various amounts of proteases in the reaction buffer (40 mM HEPES–KOH pH 7.6, 25 mM Tris–HCl pH 7.6, 4% (w/v) sucrose, 4 mM dithiothreitol, 80 μg/ml BSA, 11 mM magnesium acetate, 4 mM ATP). The reactions were incubated for 2 h at 32°C, stopped by the addition of 4× Laemmli buffer and analyzed by SDS–PAGE followed by Coomasie brilliant blue staining. To determine the amount of TrfA in each reaction, densitometric analysis was applied with Gel-Doc 2000 Imaging System (Bio-Rad) and Scion Image software (Scion Corp).
In vivo protein stability
The in vivo TrfA stability tests were performed as previously described (13). Strains used for the assay were E. coli C600 and its protease-deficient derivatives SG12050 (clpP−), ATC12017 (lon−) carrying pAT30 vectors for overproduction of wt TrfA, TrfA G254D/S267L or TrfA P151S/G254D/S267L.
Plasmid stability tests
Plasmid stability tests were performed essentially as described (28). To assay the stability of RK2 minireplicon in protease-deficient bacteria the cells of E. coli strains C600, SG12050 and ATC12017 carrying pRR10 plasmid were grown in LB medium supplemented with ampicillin (100 µg/ml) at 30°C to OD600 = 0.5. The cells were then diluted into medium without antibiotics and kept in the exponential phase of growth by sequential dilutions for at least 100 generations. To assay the stability of RK2 minireplicon in Lon-overproducing bacteria the cells of E. coli carrying pTJS42 (tetracycline resistance) and either pBAD24 or pBADLon (ampicillin resistance) plasmids were grown in LB medium supplemented with tetracycline and ampicillin at 30°C to OD600 = 0.5. The cells were then diluted into medium with only ampicillin and kept in the exponential phase of growth for no less than 40 generations. Medium was supplemented with 0.002% arabinose to induce lon expression. The rates of plasmid loss per generation were calculated as previously described.
Surface plasmon resonance analysis
Standard surface plasmon resonance (SPR) analyses using BIAcore 2000 were performed essentially as described in the manufacturer's manual. DNA binding by TrfA variants was studied using biotinylated, double-stranded DNA fragment containing five RK2 iterons immobilized on a streptavidin matrix-coated Sensor Chip SA. Running buffer used was HBS-EP (150 mM NaCl, 10 mM HEPES pH 7.4, 3 mM EDTA, 0.005% Surfactant P20). In all experiments buffer flow was set to 15 µl/min and all injections had a volume of 30 µl. The results are presented as sensorgrams obtained after subtraction of the background response signal from control experiments with buffer injections.
Size exclusion chromatography
To analyze the formation of a nucleoprotein complex, a column gel filtration method with Sepharose CL-4B was used. The reaction mixture (100 µl) included 6 µg of one of the following: ClpX, ClpA, ClpY or Lon and, if indicated, 2 µg of supercoiled plasmid DNA and 6 µg of wt TrfA in the TrfA proteolysis reaction buffer. The mixtures were incubated for 2 min at 32°C, applied on the CL-4B column (0.5 × 9 cm) and run in a column buffer (40 mM HEPES–KOH pH 7.6, 40 mM potassium glutamate, 4% (w/v) sucrose, 4 mM dithiothreitol, 10 mM magnesium acetate, 2 mM ATP, 0.01% Brij-58). Two-drop (80 µl) fractions were collected and 40 µl samples from these fractions were analyzed by SDS–PAGE followed by silver staining. 10 µl samples from the fractions were run on agarose gel with ethidium bromide to visualize DNA.
Sucrose gradient fractionation
About 50 µl reactions containing 200 ng of Alexa Fluor (Invitrogen) labeled DNA (PCR-amplified 350-bp long fragment corresponding to RK2 oriV) and/or 6 mg of either ClpA or Lon in proteolysis buffer were incubated at 32°C for 15 min and applied on the top of a 15–40% sucrose gradient prepared by the rapid freezing of 0.5 ml layers of buffer (20 mM Tris–HCl pH 7.6, 2 mM EDTA, 7 mM b-mercaptoethanol, 1 mM spermidine) containing 40, 35, 30, 25, 20 and 15% sucrose. Gradients were then centrifuged in SW 60 Ti Beckman rotor for 2 h at 46 000 r.p.m. and fractionated. Fluorescently labeled DNA was detected in the collected fractions using DTX880 Multimode Reader (Beckman-Coulter). The amount of protein in the fractions was estimated by SDS–PAGE electrophoresis followed by silver staining and densitometric analysis.
ELISA
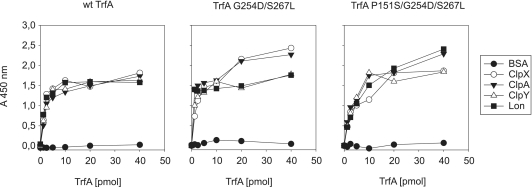
Binding of Clp chaperones and Lon to TrfA variants was analyzed by ELISA as described previously (13). Ten picomoles of BSA (negative control), ClpX, ClpA, ClpY and Lon were immobilized on the ELISA plate (Costar). Increasing amounts (1.25, 2.5, 5, 10, 20 and 40 pmol) of wt TrfA, TrfA G254D/S267L or TrfA P151S/G254D/S267L were then incubated with the immobilized proteins, unbound proteins were washed away and the relative amount of bound TrfA was detected using immunoenzymatic assay with anti-TrfA antibodies.
ATPase assay
The ATPase activity of ClpX, ClpA, ClpY and Lon was measured using a coupled enzymatic assay. Experiments were performed essentially as previously described (29) with the following modifications: buffer used was 40 mM HEPES–KOH pH 7.6, 25 mM Tris–HCl, pH 7.6, 4% (w/v) sucrose, 4 mM dithiothreitol, 80 μg/ml BSA, 11 mM magnesium acetate and no denatured luciferase was added to the mixtures. Concentrations of ATPases were: 200 nM ClpX, 180 nM ClpA, 1200 nM ClpY, 700 nM Lon and, where indicated, wt TrfA (857 nM) or supercoiled pBK20 plasmid DNA (2 µg) were added. The ATPase activity was estimated from the slope of dA340/dt curve as previously described (29).
RESULTS
TrfA proteolysis by AAA+ proteases in vitro
To address the question if and how Rep interaction with DNA would affect protein processing by proteases we purified all E. coli cytosolic AAA+ proteases and conducted a variety of in vitro experiments with TrfA replication initiator of RK2 plasmid. The obtained preparations of the E. coli ClpX, ClpA and ClpY ATPases and the ClpP and ClpQ peptidase subunits, as well as E. coli Lon protease were initially analyzed for their specific activity toward α-casein or λO protein (see ‘Materials and Methods’ section). The results showed that specific activities of purified enzymes were comparable and varied in a range from 17 to 32 U/mg.
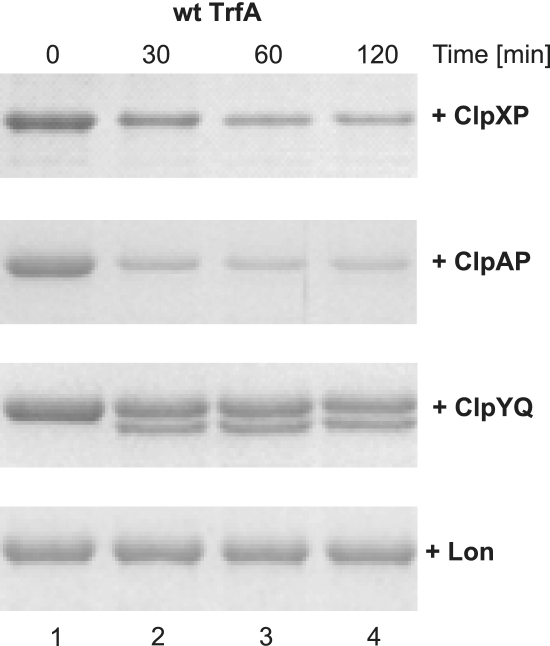
We then tested the stability of wild-type, primarily dimeric TrfA during incubation with the purified AAA+ proteases. As a result of the synthesis of a single transcript containing two alternative in-frame start codons, expression of wild-type trfA results in the synthesis of two forms of the TrfA protein: TrfA-33 and TrfA-44. Although the 44 kDa TrfA protein is required in Pseudomonas aeruginosa, the 33 kDa TrfA is sufficient for the RK2 plasmid replication in E. coli (18). Thus, during the course of this study only 33 kDa TrfA variants were used. Samples taken during the TrfA in vitro stability tests with E. coli AAA+ proteases were analyzed electrophoretically (Figure 1). When wild-type TrfA was incubated with ClpXP, we observed substantial protein degradation. This result was expected and similar to the one described previously (13). TrfA incubation with ClpAP resulted in even a more efficient proteolysis, showing that ClpAP is also able to degrade TrfA (Figure 1). The results of experiments on TrfA stability carried out with ClpYQ also demonstrated TrfA degradation, however they differed from those with ClpXP and ClpAP as we observed the appearance of a specific electrophoretic band corresponding to a lower molecular mass protein. When the TrfA protein was omitted from the reaction mixture, the lower molecular mass band was not detected (data not shown) indicating that its presence was a result of TrfA degradation by ClpYQ. Our in vitro tests with Lon protease revealed that under conditions used in the experiment the incubation of the wild-type TrfA with Lon does not result in TrfA protein degradation.
Figure 1.
TrfA in vitro proteolysis by E. coli ATP-dependent proteases. Wt TrfA protein (1.5 µg) was incubated in the reaction buffer (total mixture volume of 25 µl) for 2 h at 32°C with ClpXP (1.5 µg ClpX and 1.5 µg ClpP), ClpAP (1.5 µg ClpA and 1.5 µg ClpP), ClpYQ (1.5 µg ClpY and 1.5 µg ClpQ) or Lon (1.5 µg). Reactions were stopped at indicated times by the addition of 8 µl of 4× Laemmli buffer. Samples of 11 µl were run on SDS–PAGE and visualized by Coomasie staining.
The presence of DNA and protein oligomeric state affect TrfA proteolysis
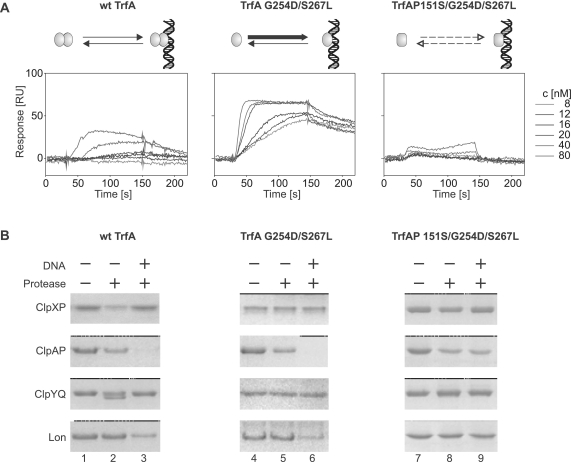
To test whether or not TrfA quaternary structure and the protein ability to bind DNA affect its processing by the E. coli AAA+ proteases, we performed experiments utilizing different TrfA variants. Wild-type TrfA remains as a dimer in the micromolar concentrations used during our in vitro proteolysis studies (20). In contrast, a previously described TrfA mutant with G254D/S267L substitutions, located within the predicted TrfA dimer interface, does not dimerize and as a monomer is hyperactive in DNA replication initiation (13,30). We also purified monomeric TrfA G254D/S267L with the additional substitution P151S which results in a protein unable to bind DNA (30,31). The isothermal circular dichroism spectra did not indicate substantial differences in the secondary structure of the TrfA variants (Supplementary Figure S1A). Secondary structure content analysis of TrfA G254D/S267L and TrfA P151S/G254D/S267L were very similar and exhibited some differences in α-helix and β-sheet content compared to wild-type TrfA (Supplementary Figure S1B). Moreover, the results of chemical denaturation studies and tryptophan fluorescence spectroscopy of the TrfA variants did not reveal any differences between TrfA P151S/G254D/S267L and replicationally active TrfA G254D/S267L monomers (Supplementary Figure S1C and D). During the chemical denaturation dimeric wild-type TrfA was slightly more stable. Light scattering studies of TrfA variants' preparations yielded similar results for all tested proteins which ruled out the possibility of any of the proteins being more prone to aggregation (Supplementary Figure S1E). Using Surface Plasmon Resonance (SPR) (‘Materials and Methods’ section) we analyzed the interaction of wild-type TrfA and the two TrfA mutant protein variants with a 129 bp linear double-stranded DNA fragment containing the sequence of five iterons present in RK2 oriV. Real-time kinetics experiments demonstrated significant differences among the analyzed TrfA proteins in their ability to form complexes with DNA (Figure 2A). Whereas TrfA G254D/S267L interacted with DNA immobilized on a sensor chip with high efficiency, TrfA P151S/G254D/S267L nucleoprotein complex formation was severely impaired. Under the same experimental conditions, wild-type TrfA interacted with DNA, though not as efficiently as was observed for the TrfA G254D/S267L variant. To test if the E. coli AAA+ proteases were able to degrade the analyzed TrfA protein variants, in vitro proteolytic reactions were performed in the absence and presence of iterons-containing plasmid pBK20 supercoiled DNA (Figure 2B). As we observed in the previous experiment (Figure 1), ClpXP was able to degrade wild-type TrfA in the absence of plasmid DNA; however, the addition of DNA to the reaction mixture inhibited the proteolytic reaction (Figure 2B, compare lanes 2 and 3). Both TrfA monomeric mutants, TrfA G254D/S267L and TrfA P151S/G254D/S267L, were resistant to ClpXP degradation, regardless of the presence or absence of DNA in the reaction mixture. In contrast, in assays with ClpAP, all three TrfA variants were degraded. Interestingly, we found that DNA substantially stimulated ClpAP-dependent degradation of the dimeric wild-type TrfA and the TrfA G254D/S267L monomeric mutant (Figure 2B, lanes 2, 3, 5, 6). The stimulation of proteolysis by DNA was not observed for the TrfA P151S/G254D/S267L monomer, which does not interact with DNA (Figure 2B, lanes 8 and 9).The ClpYQ protease did not degrade either of the TrfA monomeric variants, regardless of DNA presence (Figure 2B, lanes 5, 6, 8, 9) and as was observed for ClpXP, we found that wild-type TrfA proteolysis by ClpYQ was inhibited by the addition of DNA (Figure 2B, lanes 2 and 3). The addition of DNA to the reaction mixtures revealed Lon activity toward TrfA as a substrate. Wild-type TrfA and the TrfA G254D/S267L monomeric variant were degraded by Lon only in the presence of DNA (Figure 2B, lanes 3 and 6). We did not observe any Lon-dependent proteolysis of TrfA P151S/G254D/S267L, the mutant protein which is not able to bind DNA (Figure 2B, lanes 8 and 9). It must be pointed out that DNA effects observed during our experiments were similar when instead of pBK20 plasmid we used pUC19 which does not contain oriV iterons (data not shown). This was probably due to non-specific TrfA interaction with pUC19 DNA. However, we could not exclude that also proteases bound DNA and this interaction affected their activity.
Figure 2.
DNA affects TrfA in vitro proteolysis. Panel (A) shows the results of experiments in which DNA binding was tested for the dimeric wt TrfA and the monomeric mutants TrfA G254D/S267L and TrfA P151S/G254D/S267L. Sensorgrams show the SPR analysis results of binding of each of the TrfA variants to a double-stranded DNA fragment containing five RK2 iterons (frag1). Injections contained the indicated concentrations of TrfA variants in HBS-EP buffer. HBS-EP was also used as a running buffer. Panel (B) shows the results of SDS–PAGE analysis of TrfA variants in vitro proteolysis reactions with or without DNA in the reaction mixture. Proteolysis was carried out as described in Figure 1 with the following modifications: proteolysis of wt TrfA by ClpAP was performed using 150 ng ClpA and 500 ng ClpP; volumes of the reaction mixtures for TrfA G254D/S267L proteolysis were 50 µl; reactions in lanes 3, 6 and 9 contained 500 ng of supercoiled pBK20 plasmid DNA.
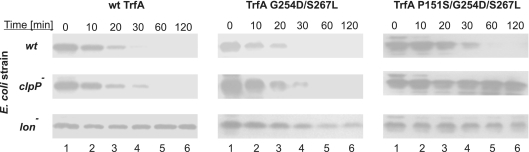
Since our in vitro experiments exposed substantial differences in degradation of the TrfA mutants by the E. coli AAA+ proteases, we decided to test the stability of these mutants in vivo. Tests based on translation inhibition (see ‘Materials and Methods’ section) were performed in wild-type E. coli and isogenic strains having mutations in either the clpP gene or the lon gene. The results demonstrate that all three TrfA variants were efficiently proteolysed in wild-type E. coli cells (Figure 3). The stabilities of wild-type TrfA and TrfA G254D/S267L slightly increased and the stability of TrfA P151S/G254D/S267L significantly increased in the E. coli clpP− mutant, indicating the role of ClpXP and/or ClpAP in the degradation of these proteins. Substantial stabilization of all the analyzed proteins was observed in the E. coli lon− strain showing that Lon protease might have a major impact on the TrfA stability in vivo. Moreover, and consistent with our in vitro proteolytic tests, the TrfA P151S/G254D/S267L mutant was the most stable of the tested protein variants in vivo.
Figure 3.
In vivo stability of TrfA variants in wild-type and protease-deficient strains. The stability of TrfA protein was analyzed after inhibition of translation by tetracycline in TrfA-overproducing bacteria. Cells of E. coli strains C600, C600 clpP− and C600 lon− harboring plasmids for overproduction of wt TrfA, TrfA G254D/S267L or TrfA P151S/G254D/S267L were used. Samples taken from the cultures at indicated time points after the addition of tetracycline were analyzed for TrfA presence by SDS–PAGE followed by immunoblot with anti-TrfA antibodies.
RK2 minireplicon stability in protease-deficient or protease-overproducing E. coli cells
To test how the absence of proteases affects plasmid replication in vivo we performed RK2 minireplicon stability tests using wild-type and protease-deficient E. coli strains. The replication of mini-RK2 plasmids, which contain only RK2 origin of replication, trfA gene and an antibiotic resistance gene, is utterly dependent on TrfA provided in cis. Therefore, we used mini-RK2 derivative, plasmid pRR10, carrying ampicillin resistance, in order to estimate the effects of protease absence on the TrfA-mediated plasmid replication. Although the E. coli strains with specific mutations in genes encoding proteases were transformed with pRR10 with similar efficiencies as the wild-type strain, we observed differences in the stability of pRR10 plasmid. The results of the analysis are shown in Table 1. Comparing to the wild-type strain the rate of plasmid loss was two times higher in clpP− strain and almost three times higher in lon− strain.
Table 1.
Stability of RK2 minireplicons in protease deficient and Lon-overproducing strains
| Escherichia coli C600 strain | Plasmid loss rate (% per generation)a |
|---|---|
| Wild-typeb | 0.12 ± 0.01 |
| clpP−b | 0.24 ± 0.02 |
| lon−b | 0.33 ± 0.01 |
| Wild-type (pBAD24)c | 1.46 ± 0.05 |
| Wild-type (pBADLon)c | 3.23 ± 0.94 |
aGiven values are means from three independent repeats of each experiment.
bStability of pRR10 minireplicon was analyzed in C600 and its mutant derivatives cultured in antibiotic-free medium
cStability of pTJS42 minireplicon was analyzed in wild-type C600 strain cells carrying either pBAD24 or pBADLon cultured in ampicillin-containing medium.
Because Lon protease appeared to have the strongest effect on the in vivo stability of the TrfA protein, as well as the mini-RK2 plasmid stability, we decided to test the stability of mini-RK2 derivative in Lon-overproducing cells. We performed stability tests of the pTJS42 plasmid, which contains oriV, trfA gene and tetracycline resistance gene, in cells of E. coli C600 strain carrying either a plasmid with arabinose-inducible lon gene (pBADLon) or an empty vector (pBAD24) (Table 1). During the course of the experiment the cells were grown in ampicillin-containing medium to maintain the pBAD plasmids. Although for both types of cells the rates of mini-RK2 plasmid loss per generation were relatively high, most likely because of the ampicillin resistance selection, the value was over two times higher for Lon-overproducing cells.
Interaction of the AAA+ proteases with DNA and TrfA
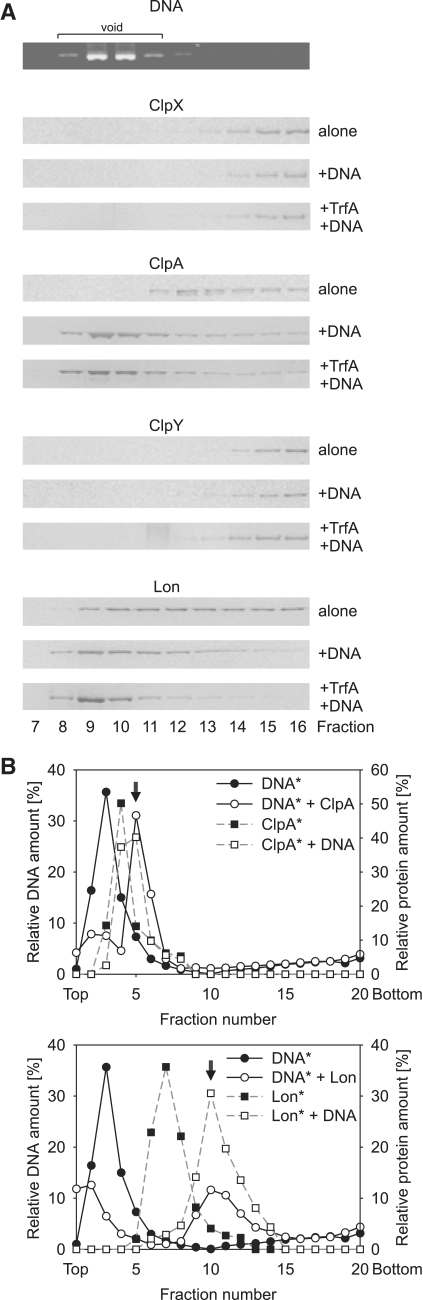
Because our in vitro experiments showed that DNA was a factor affecting TrfA degradation by E. coli AAA+ proteases, we asked if proteases could interact with DNA or with DNA bound by the TrfA protein. To answer this question, we applied size exclusion chromatography using a Sepharose CL-4B column (see ‘Materials and Methods’ section). Experiments were performed with ClpX, ClpA, ClpY and Lon (Figure 4A). In a control experiment, neither DNA nor TrfA were present in the reaction mixtures. After the addition of supercoiled plasmid pBK20 DNA or plasmid pBK20 DNA bound by the wild-type TrfA, substantial amounts of ClpA and Lon were detected in the column void volume, indicating that both proteins are able to form nucleoprotein complexes, regardless of the presence or absence of the substrate protein. Under the same experimental conditions, we did not observe ClpX or ClpY (Figure 4A) as well as ClpP or ClpQ (data not shown) in the void fractions containing plasmid DNA. DNA binding by E. coli and eukaryotic mitochondrial Lon proteins has been previously reported (32–35); however, the E. coli ClpA interaction with DNA was unexpected. We applied the sucrose gradient fractionation to confirm that ClpA and Lon are able to form nucleoprotein complexes with linear DNA. Fluorescently labeled 350-bp long DNA fragment was used in the experiment. The results showed that the positions of DNA fragment and the proteins move toward the bottom fractions of the gradient when two components are incubated together, comparing to incubation of either component alone (Figure 4B), although the effect was stronger for Lon. This indicates that both ClpA and Lon are indeed capable of formation of nucleoprotein complex not only with supercoiled, but also shorter, linear DNA fragment. Using SPR, we confirmed that result with a 129 bp pUC19 dsDNA fragment or 129 bp long iterons-containing dsDNA fragment. The results showed that both proteins can interact with analyzed DNA fragments (Supplementary Figure S2). To further characterize these interactions, we applied electrophoretic mobility shift assay with a 135 and 350 bp-long DNA probes. Presence of ClpA or Lon resulted with mobility shift of analyzed DNA fragments; however, the addition of a non-specific competitor [poly d(I-C)] decreased the observed effects (Supplementary Figure S3). This suggests that the interactions are non-specific rather than limited to some defined sequence present within the used DNA fragments. In contrast, in the control reaction with TrfA G254D/S267L discrete bands appear upon the addition of poly d(I-C), indicating high specificity of binding to iterons.
Figure 4.
Nucleoprotein complex formation by Clp and Lon proteins. (A) Nucleoprotein complex formation was studied using gel filtration. Reaction mixtures containing ClpX, ClpA, ClpY or Lon were incubated for 2 min at 32°C alone, in the presence of pBK20 plasmid DNA or in the presence of pBK20 and wt TrfA. After the pre-incubation step mixtures were run through a CL-4B column in the column buffer. Collected fractions were analyzed by SDS–PAGE and silver staining for the presence of proteins and by agarose electrophoresis for the presence of DNA. Plasmid DNA was found in the same fractions in all experiments (top panel). (B) Sucrose gradient fractionation of nucleoprotein complexes formed by ClpA and Lon with 350 bp-long DNA fragment was performed as described in ‘Materials and Methods’ section. Reactions contained DNA alone, protein alone or both components. Graphs show the amount of DNA and protein in the collected fractions. Asterisks in the graph legend indicate which component was detected. Arrows indicate the positions of nucleoprotein complexes.
The obtained data describe Lon and ClpA interactions with DNA. We also decided to analyze the AAA+ proteases interactions with TrfA by ELISA. We studied the binding of three TrfA variants to immobilized proteins ClpX, ClpA, ClpY, Lon and BSA (Figure 5) and detected interactions of each TrfA variant with all proteins tested except BSA (Figure 5). Interestingly, ClpX and ClpY interact with the TrfA monomeric mutants TrfA G254D/S267L and TrfA P151S/G254D/S267L, despite the fact that these variants were not degraded by ClpXP and ClpYQ (Figure 2B). To confirm these results, we performed SPR on a CM5 sensor chip with the wild-type TrfA, TrfA G254D/S267L and TrfA P151S/G254D/S267L immobilized on the surface (Supplementary Figure S4). Injections of ClpX, ClpA, ClpY and Lon resulted with increase in the response signal indicating efficient interactions between proteins and TrfA variants tested.
Figure 5.
Interactions of Clp and Lon proteins with TrfA variants. Binding of TrfA variants to Clp chaperones and Lon was analyzed by ELISA as described under ‘Materials and Methods’ section. BSA protein was used as a negative control.
Effects of DNA and substrate on ATPase activity of proteases
Our experiments showed that despite the fact that DNA was not mandatory for TrfA interaction with the E. coli AAA+ proteases, it did affect proteases' activity in TrfA degradation. While Lon and ClpAP proteolytic activities were stimulated, the proteolytic activities of ClpXP and ClpYQ were inhibited by the presence of DNA. To explore the mechanism(s) for these opposing effects, we asked if and how the ATPase activity of the analyzed enzymes would be influenced by DNA and the substrate protein. The ATPase activity tests were performed using an enzymatic coupled assay (see ‘Materials and Methods’ section) and standardized using each ATPase's activity alone as 100%. The mixtures contained ATPase (ClpX, ClpA, ClpY or Lon) with TrfA protein or plasmid pBK20 supercoiled DNA or with both components. We observed that plasmid DNA slightly stimulated ATPase activity of ClpA and Lon, while it had no effect on the activity of ClpX and ClpY (Table 2). The ATPase stimulation of ClpA and Lon was even more pronounced when both DNA and the wild-type TrfA were present in the reaction mixture (Table 2). The addition of the wild-type TrfA alone caused minor inhibition of the ATPase activity of all analyzed enzymes. This inhibition of baseline activity may be due to slow processing of the substrate protein by the tested ATPases.
Table 2.
Effect of TrfA and DNA on ATPase activity of Clp chaperones and Lon
| ATPase | ATPase activity (%)a |
||
|---|---|---|---|
| + wt TrfA | + DNA | + wt TrfA, DNA | |
| ClpX | 83.1 ± 3.4 | 90.3 ± 6.8 | 92.8 ± 3.4 |
| ClpA | 84.1 ± 7.3 | 127.7 ± 12.4 | 143.2 ± 10.6 |
| ClpY | 89.9 ± 2.4 | 91.9 ± 3.3 | 85.2 ± 2.4 |
| Lon | 78.0 ± 15.6 | 104.2 ± 7.7 | 181.5 ± 30.4 |
aGiven values are means from three independent repeats of each experiment. Results were normalized to the activity of each ATPase alone.
TrfA proteolysis stimulation by DNA depends on the order of addition of reaction components
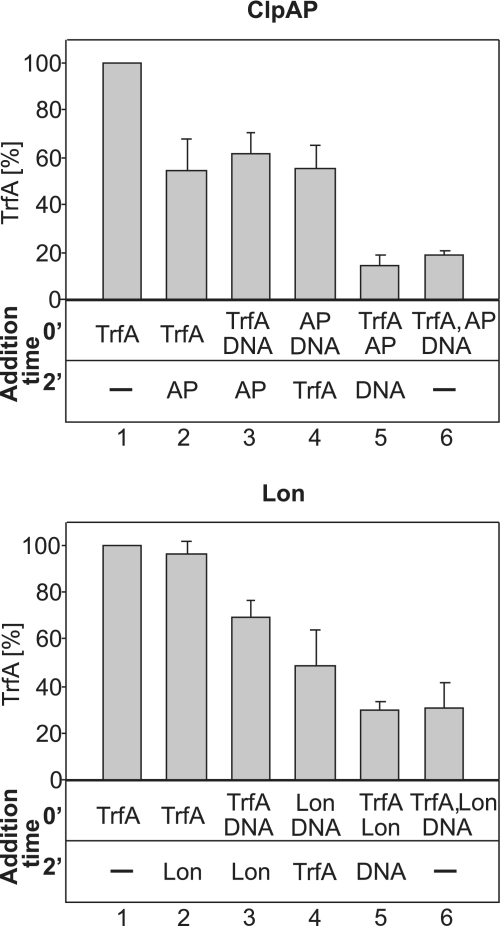
To further analyze the mechanism(s) of how DNA affects the proteases activity, we designed experiments in which the order of addition of the reaction components varied. The reactions were designed to test if pre-formation of complexes consisting of protease, TrfA and DNA could stimulate or inhibit subsequent proteolytic reaction. The results showed that ClpAP proteolytic activity was highest when the TrfA protein was pre-incubated with ClpAP for 2 min before the addition of DNA or when TrfA, the protease and the DNA were added at the same time (Figure 6 top panel). As we observed in our previous experiments (Figure 2B), Lon proteolytic activity toward TrfA was detected only in the presence of DNA (Figure 6 lower panel). Regardless of the addition order, when DNA was present in the reaction mixture, we observed TrfA degradation by Lon. The most efficient degradation occurred when TrfA was pre-incubated for 2 min with Lon before the addition of DNA, or when all three reaction components, TrfA, Lon and DNA, were added at the same time. These results suggest that both ClpAP and Lon, when complexed with the substrate protein, can interact with DNA, and this interaction most probably stimulates the proteolytic reaction. Using variable addition order of reaction components, we also conducted proteolytic tests with ClpXP and ClpYQ (Supplementary Figure S5). The results showed that regardless of the order of addition of the reaction components, DNA had a similar inhibitory effect on both proteases.
Figure 6.
Addition order of reaction components affects the efficiency of TrfA degradation by ClpAP and Lon in vitro. TrfA proteolysis reactions were performed essentially as described in ‘Materials and Methods’ section. The volumes of reaction mixtures were 25 µl. Reaction components—wt TrfA, pBK20 and protease (ClpAP or Lon)—were mixed together in different orders. For reactions 3–5, the mixtures containing two components were initially pre-incubated for 2 min at 32°C, then the remaining component was added. For reactions 1 and 6, there was no pre-incubation step. All reaction mixtures were further incubated for 2 h at 32°C and analyzed by SDS–PAGE, followed by Coomasie staining and densitometric analysis of the amount of TrfA. All bars on the graphs represent mean results of three independent experiments. Amounts of proteases used were 150 ng ClpA and 500 ng ClpP or 1.5 µg Lon.
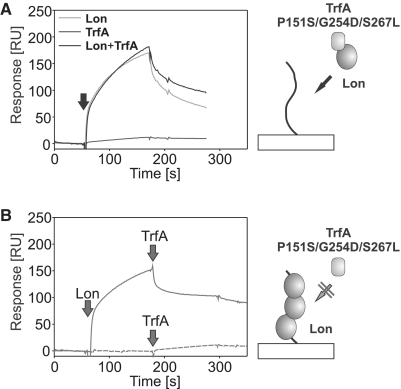
The analysis of Lon–TrfA–DNA nucleoprotein complex formation
The TrfA variant with substitutions P151S/G254D/S267L, which we showed to interact with both ClpA and Lon (Figure 5 and Supplementary Figure S4) but was DNA binding deficient (Figure 2A), allowed us to further analyze the interaction of the proteases with DNA. To test the possibility that Lon could interact with DNA in the substrate-bound state, we applied the SPR method allowing for the real-time analysis. Unfortunately, due to limitations of the SPR method, we were not able to find appropriate buffer conditions to study the interaction of the ClpA–TrfA complex with DNA. Thus, experiments were performed only with Lon and the TrfA P151S/G254D/S267L mutant. It must be noted that TrfA P151S/G254D/S267L while being able to be bound by Lon, was not degraded by this protease (Figure 2B). The protease and the substrate, together or alone, were incubated prior to injection onto a sensor chip with the immobilized pUC19 dsDNA fragment. In control experiments, Lon alone gave a response signal similar to the preformed Lon–TrfA complex, while the TrfA mutant alone did not interact with the dsDNA fragment efficiently (Figure 7A). These results showed that when the protease forms a complex with TrfA, it retains the DNA binding ability. We also tested if the TrfA mutant protein was able to interact with Lon bound to DNA (Figure 7B). Lon injection onto the sensor chip containing the pUC19 dsDNA fragment increased the response signal, indicating Lon–DNA interaction. Subsequent injection of TrfA P151S/G254D/S267L did not result in a further response signal increase. These results demonstrated that the TrfA mutant protein is not able to interact with Lon when the protease is bound to DNA. It is consistent with the order of addition experiment results obtained for Lon (Figure 6) and explains why we observed only partial proteolysis stimulation after initial Lon preincubation with DNA.
Figure 7.
Formation of Lon–TrfA–DNA nucleoprotein complex. Formation of nucleoprotein complex involving Lon and TrfA in real-time was studied using SPR on a sensor chip with immobilized pUC19 DNA fragments (frag2) on its surface. Buffer used for the analysis was HBS-EP supplemented with 10 mM magnesium acetate and 2 mM ATP. (A) Lon (200 nM), TrfA P151S/G254D/S267L (80 nM) or both were pre-incubated for 2 min in HBS-EP and subsequently injected onto sensor chip. (B) Injection of 200 nM Lon (solid line) or buffer (dashed line) onto sensor chip was rapidly followed by injection of 80 nM TrfA P151S/G254D/S267L.
DISCUSSION
TrfA protein is a universal substrate for AAA+ proteases
In the present study, we demonstrate that the TrfA protein is recognized and processed essentially by all E. coli AAA+ cytoplasmic proteases including ClpXP, ClpAP, ClpYQ and Lon. It was observed previously that proteases have overlapping specificities toward some substrates (36,37). However, we show for the first time that all E. coli cytosolic proteases are capable of degradation of a single, specific substrate, which is a key player in plasmid replication. During our experiments, the observed TrfA proteolysis by the ClpYQ protease was incomplete and resulted in the partial protein degradation. Most likely TrfA is released by the protease after partial proteolysis. Similar degradation product was also observed during TrfA processing by ClpXP (13). In contrast to ClpXP and ClpYQ, the degradation of TrfA by ClpAP and Lon was more efficient. According to the in vivo protein stability tests, Lon appears to be the main cellular protease responsible for TrfA proteolysis.
TrfA quaternary structure affects its proteolysis by ClpXP and ClpYQ but not by ClpAP and Lon
Experiments presented in this work show that TrfA oligomeric state affects its proteolysis by both ClpXP and ClpYQ, which are able to degrade TrfA dimers only. Previously, we demonstrated that ClpXP degrades TrfA both in vivo and in vitro, and this proteolysis was also limited to the dimeric form of the protein (13). Interestingly, current data shows that both ClpX and ClpY interact with TrfA dimers and monomers indicating that the proteases' inability to degrade monomeric TrfA is not a result of an inability to recognize it. Changes in quaternary structure of the target protein were proposed to affect degrons accessibility and therefore the efficiency of proteolysis (7). For example, it was demonstrated that the UvrA protein is protected from degradation upon its interaction with UvrB (38). It was also shown that UmuD’ is a substrate for the ClpXP protease, but for the efficient proteolysis to commence, it must form a heterodimer with its precursor, UmuD (39). Our results show that although the changes in quaternary structure do not substantially affect substrate recognition, they may dramatically affect substrate processing by AAA+ protease. In contrast to ClpXP and ClpYQ, ClpAP and Lon are able to fully degrade TrfA dimers and monomers with similar efficiencies indicating that these proteases are not sensitive to changes in TrfA oligomeric state.
DNA as a factor inhibiting TrfA proteolysis by ClpXP and ClpYQ
Our experiments revealed that DNA tremendously affects the activity of E. coli AAA+ proteases in the degradation of TrfA protein. DNA presence in the reaction inhibits ClpXP and ClpYQ proteolytic activity toward TrfA, yet has no effect on the ATPase activity of these proteases' unfoldase units. The inhibition of proteolytic activity was observed regardless of the order of addition of reaction components. Since we show that ClpX and ClpY (as well as ClpP and ClpQ) do not interact with DNA, the observed inhibition of TrfA degradation by DNA must be a result of TrfA interaction with DNA. Our ELISA and SPR results clearly demonstrate that TrfA is recognized and can be bound by ClpX and ClpY ATPases. However, when the TrfA protein binds DNA, it is no longer recognized either by ClpX or ClpY, as we did not observe their interaction with DNA–TrfA complex in the size exclusion chromatography. Consistently with our observations on TrfA, it was shown that binding of λO protein to ori lambda DNA protects it from degradation by ClpXP (40). Similar effects have been reported for SoxS and ZntR proteins (41,42). Another example of the altered specificity of substrate recognition upon formation of a nucleoprotein complex is the remodeling of the bacteriophage Mu strand transfer complex. Although the Mu strand transfer complex contains a MuA tetramer, in vitro experiments indicate that ClpX unfolds only one MuA subunit (43).
DNA as a factor stimulating TrfA proteolysis by ClpAP and Lon
While DNA presence in the reaction inhibits ClpXP and ClpYQ proteolytic activities toward TrfA, it stimulates TrfA degradation by ClpAP and Lon. We observed similar stimulatory effects on the proteolysis of other replication initiators: RepE of plasmid F and λO of bacteriophage λ (S. Kubik. and I. Konieczny, unpublished data), which suggests that the phenomenon might be common for this class of protease substrates. Here, we performed experiments with TrfA to elucidate the mechanism underlying this stimulation. It must be pointed out that our results clearly demonstrate that ClpAP alone can degrade TrfA, and DNA only enhances the extent of this degradation. In contrast, TrfA degradation by Lon is strictly DNA dependent. Similar strict dependency was reported for polyphosphate-induced ribosomal proteins degradation by Lon (44,45). Our data obtained from size exclusion chromatography, sucrose gradient fractionation, SPR and EMSA demonstrate that, in contrast to ClpX and ClpY, ClpA and Lon can interact with both supercoiled and linear DNA. These interactions appear not to be sequence specific. DNA binding by bacterial, as well as eukaryotic mitochondrial Lon proteins has been previously reported (32–35), however the E. coli ClpA interaction with DNA was unexpected. Although the structure and functions of Lon have been studied extensively the exact location of the DNA binding domain is still unknown (45–47). ClpA does not contain any well known DNA binding motifs. By utilizing bioinformatic approach with different algorithms, we found two particular regions of ClpA that have a potential to interact with DNA (S. Kubik. and I. Konieczny, unpublished data). However, in ClpA hexamer (48), both motifs are not exposed to the surface; therefore, their involvement in the process of DNA binding and proteolysis stimulation is arguable. Similar approach did not yield results for Lon.
It could be considered that the ClpA and Lon interactions with DNA stimulate the enzymes ATPase activities, which might result in a more efficient proteolysis. In our experiments, the ATPase activities of Lon and ClpA were only slightly stimulated by DNA alone; however, the presence of both DNA and TrfA substantially increased ATP hydrolysis of both enzymes. This result ruled out the possibility that DNA simply accelerates the turnover of proteases' unfoldase units and it is consistent with previous reports showing that the ATPase activity of Lon was enhanced by DNA in the presence of casein (49,50).
Our data demonstrate that both ClpA and Lon are able to interact with DNA, regardless of the presence of the substrate protein (Figure 4). Since TrfA also binds DNA, it makes the analysis of the mechanism of proteolysis stimulation by DNA very complex. Identification of the nature and the kinetics of the interactions is a key to understanding the stimulatory effect of DNA on the proteases activities. ClpA and Lon can bind the wild-type TrfA and the analyzed TrfA mutant proteins in the absence of DNA. This indicates that the substrate protein interaction with DNA is not needed for its recognition by ClpA and Lon. Interestingly, the lack of the proteolysis stimulation with the DNA-binding defective mutant TrfA P151S/G254D/S267L suggests the importance of substrate–DNA interaction for its degradation. As tested in this work, TrfA P151S/G254D/S267L is more stable in vivo than DNA-binding variants of the protein. Interestingly, the Lon-dependent degradation of casein and globin, non-physiological Lon substrates that do not form nucleoprotein complexes, is also stimulated by the addition of DNA (49,50). Taken together these results indicate that Lon activity could be stimulated by the protease direct interaction with DNA or both the protease and substrate interactions with DNA.
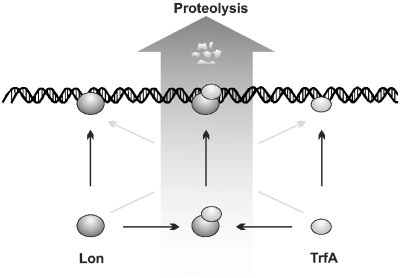
Our data show that ClpA and Lon bind the substrate protein and DNA. Therefore, when TrfA forms a nucleoprotein complex, the protease can bind DNA at a location distant from such a complex, which decreases the possibility of its interaction with TrfA. This could explain the inefficient DNA stimulatory effect during our experiments when TrfA was incubated with DNA prior to the addition of the protease (Figure 6). The same mechanism could explain the fact that only a slight stimulation (Lon) or no stimulation at all (ClpAP) was observed when the protease–DNA complex was formed before TrfA addition. The extent of proteolysis stimulation by DNA was the highest when TrfA and ClpAP or Lon were pre-incubated prior to the addition of DNA or when all components were added simultaneously. Interestingly, the SPR experiments with TrfA P151S/G254D/S267L show that the formation of the Lon–TrfA complex does not inhibit Lon interaction with DNA but when the protease forms a complex with DNA as it is not able to further interact with TrfA (Figure 7). It has been suggested earlier that Lon may become ‘entrapped’ by DNA to prevent uncontrolled proteolysis (51). These data also explain weak stimulation of Lon when the protease–DNA complex was formed before TrfA addition. Our results indicate that the protease–substrate complex must be pre-formed before DNA can trigger its stimulatory effect. We cannot exclude that Lon can also interact with TrfA already bound to DNA. Clearly the degradation of TrfA by Lon can be carried out only on DNA (Figure 8). Lon was reported to co-localize with the nucleoid (52,53) and was proposed to be a possible factor responsible for degradation of various DNA-binding regulatory proteins. Our results demonstrate that indeed a DNA replication protein, when interacting with DNA, is subjected to efficient degradation by Lon protease.
Figure 8.
Model explaining the effect of DNA on TrfA proteolysis by Lon. Both Lon and TrfA possess DNA-binding ability. Lon associates with TrfA but is not able to degrade it efficiently, unless the Lon–TrfA complex binds to DNA. This binding triggers the proteolysis of substrate protein.
Although the already published data, as well as data presented in the current study, demonstrate that the TrfA protein can be processed by chaperones or proteases in vitro (8,9,13), the in vivo tests (8) did not reveal to date, if and how those proteins could affect RK2 plasmid activity in bacterial cells. The reason for this is the TrfA specificity as a universal substrate, which results in the overlapping activities of chaperones and various proteases toward this protein. In the presented work, we demonstrate that a specific host protease, namely Lon, affects the stability of RK2 derivatives which was slightly decreased by both the absence and the overproduction of Lon. Direct and indirect effects of Lon activity on the plasmid stability cannot be excluded; however, we show that Lon can degrade TrfA both in vivo and in vitro. Since TrfA proteolysis by Lon is limited to nucleoprotein complexes, the initiation complex, as well as ‘handcuff’ structure formed by TrfA on plasmid RK2 DNA, are likely candidates for Lon activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR online: Supplementary Figures S1–5.
FUNDING
Funding for open access charge: Polish Ministry of Science and Higher Education (N N301 295837); Foundation for Polish Science (TEAM/2009-3/5 to I.K. and S.K.).
Conflict of interest statement. None declared.
Supplementary Material
ACKNOWLEDGEMENTS
The authors thank Prof. Bernd Bukau and Prof. Matthias Bochtler for providing them with plasmids for overproduction of proteins. The authors are grateful to Prof. Donald Helinski and Dr Aresa Toukdarian for critically reading the manuscript.
REFERENCES
- 1.Dougan DA, Mogk A, Bukau B. Protein folding and degradation in bacteria: to degrade or not to degrade? That is the question. Cell Mol. Life Sci. 2002;59:1607–1616. doi: 10.1007/PL00012487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Tsilibaris V, Maenhaut-Michel G, Van Melderen L. Biological roles of the Lon ATP-dependent protease. Res. Microbiol. 2006;157:701–713. doi: 10.1016/j.resmic.2006.05.004. [DOI] [PubMed] [Google Scholar]
- 3.Baker TA, Sauer RT. ATP-dependent proteases of bacteria: recognition logic and operating principles. Trends Biochem. Sci. 2006;31:647–653. doi: 10.1016/j.tibs.2006.10.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Park SC, Jia B, Yang JK, Van DL, Shao YG, Han SW, Jeon YJ, Chung CH, Cheong GW. Oligomeric structure of the ATP-dependent protease La (Lon) of Escherichia coli. Mol. Cells. 2006;21:129–134. [PubMed] [Google Scholar]
- 5.Gur E, Sauer RT. Degrons in protein substrates program the speed and operating efficiency of the AAA+ Lon proteolytic machine. Proc. Natl Acad. Sci. USA. 2009;106:18503–18508. doi: 10.1073/pnas.0910392106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kirstein J, Moliere N, Dougan DA, Turgay K. Adapting the machine: adaptor proteins for Hsp100/Clp and AAA+ proteases. Nat. Rev. Microbiol. 2009;7:589–599. doi: 10.1038/nrmicro2185. [DOI] [PubMed] [Google Scholar]
- 7.Schmidt R, Bukau B, Mogk A. Principles of general and regulatory proteolysis by AAA+ proteases in Escherichia coli. Res. Microbiol. 2009;160:629–636. doi: 10.1016/j.resmic.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 8.Konieczny I, Helinski DR. The replication initiation protein of the broad-host-range plasmid RK2 is activated by the ClpX chaperone. Proc. Natl Acad. Sci. USA. 1997;94:14378–14382. doi: 10.1073/pnas.94.26.14378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Konieczny I, Liberek K. Cooperative action of Escherichia coli ClpB protein and DnaK chaperone in the activation of a replication initiation protein. J. Biol. Chem. 2002;277:18483–18488. doi: 10.1074/jbc.M107580200. [DOI] [PubMed] [Google Scholar]
- 10.Kruklitis R, Welty DJ, Nakai H. ClpX protein of Escherichia coli activates bacteriophage Mu transposase in the strand transfer complex for initiation of Mu DNA synthesis. EMBO J. 1996;15:935–944. [PMC free article] [PubMed] [Google Scholar]
- 11.Zzaman S, Reddy JM, Bastia D. The DnaK-DnaJ-GrpE chaperone system activates inert wild type pi initiator protein of R6K into a form active in replication initiation. J. Biol. Chem. 2004;279:50886–50894. doi: 10.1074/jbc.M407531200. [DOI] [PubMed] [Google Scholar]
- 12.Levchenko I, Luo L, Baker TA. Disassembly of the Mu transposase tetramer by the ClpX chaperone. Genes Dev. 1995;9:2399–2408. doi: 10.1101/gad.9.19.2399. [DOI] [PubMed] [Google Scholar]
- 13.Pierechod M, Nowak A, Saari A, Purta E, Bujnicki JM, Konieczny I. Conformation of a plasmid replication initiator protein affects its proteolysis by ClpXP system. Protein Sci. 2009;18:637–649. doi: 10.1002/pro.68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wojtkowiak D, Georgopoulos C, Zylicz M. Isolation and characterization of ClpX, a new ATP-dependent specificity component of the Clp protease of Escherichia coli. J. Biol. Chem. 1993;268:22609–22617. [PubMed] [Google Scholar]
- 15.Wickner S, Gottesman S, Skowyra D, Hoskins J, McKenney K, Maurizi MR. A molecular chaperone, ClpA, functions like DnaK and DnaJ. Proc. Natl Acad. Sci. USA. 1994;91:12218–12222. doi: 10.1073/pnas.91.25.12218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Moreno-del Alamo M, Sanchez-Gorostiaga A, Serrano AM, Prieto A, Cuellar J, Martin-Benito J, Valpuesta JM, Giraldo R. Structural analysis of the interactions between hsp70 chaperones and the yeast DNA replication protein Orc4p. J. Mol. Biol. 2010;403:24–39. doi: 10.1016/j.jmb.2010.08.022. [DOI] [PubMed] [Google Scholar]
- 17.Nishitani H, Taraviras S, Lygerou Z, Nishimoto T. The human licensing factor for DNA replication Cdt1 accumulates in G1 and is destabilized after initiation of S-phase. J. Biol. Chem. 2001;276:44905–44911. doi: 10.1074/jbc.M105406200. [DOI] [PubMed] [Google Scholar]
- 18.Kolatka K, Kubik S, Rajewska M, Konieczny I. Replication and partitioning of the broad-host-range plasmid RK2. Plasmid. 2010;64:119–134. doi: 10.1016/j.plasmid.2010.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Giraldo R. Common domains in the initiators of DNA replication in Bacteria, Archaea and Eukarya: combined structural, functional and phylogenetic perspectives. FEMS Microbiol. Rev. 2003;26:533–554. doi: 10.1111/j.1574-6976.2003.tb00629.x. [DOI] [PubMed] [Google Scholar]
- 20.Toukdarian AE, Helinski DR, Perri S. The plasmid RK2 initiation protein binds to the origin of replication as a monomer. J. Biol. Chem. 1996;271:7072–7078. doi: 10.1074/jbc.271.12.7072. [DOI] [PubMed] [Google Scholar]
- 21.Gottesman S, Gottesman M, Shaw JE, Pearson ML. Protein degradation in E. coli: the lon mutation and bacteriophage lambda N and cII protein stability. Cell. 1981;24:225–233. doi: 10.1016/0092-8674(81)90518-3. [DOI] [PubMed] [Google Scholar]
- 22.Kittell BL, Helinski DR. Iteron inhibition of plasmid RK2 replication in vitro: evidence for intermolecular coupling of replication origins as a mechanism for RK2 replication control. Proc. Natl Acad. Sci. USA. 1991;88:1389–1393. doi: 10.1073/pnas.88.4.1389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Thompson MW, Maurizi MR. Activity and specificity of Escherichia coli ClpAP protease in cleaving model peptide substrates. J. Biol. Chem. 1994;269:18201–18208. [PubMed] [Google Scholar]
- 24.Maurizi MR, Thompson MW, Singh SK, Kim SH. Endopeptidase Clp: ATP-dependent Clp protease from Escherichia coli. Methods Enzymol. 1994;244:314–331. doi: 10.1016/0076-6879(94)44025-5. [DOI] [PubMed] [Google Scholar]
- 25.Bochtler M, Hartmann C, Song HK, Bourenkov GP, Bartunik HD, Huber R. The structures of HsIU and the ATP-dependent protease HsIU-HsIV. Nature. 2000;403:800–805. doi: 10.1038/35001629. [DOI] [PubMed] [Google Scholar]
- 26.Bochtler M, Ditzel L, Groll M, Huber R. Crystal structure of heat shock locus V (HslV) from Escherichia coli. Proc. Natl Acad. Sci. USA. 1997;94:6070–6074. doi: 10.1073/pnas.94.12.6070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Goldberg AL, Moerschell RP, Chung CH, Maurizi MR. ATP-dependent protease La (lon) from Escherichia coli. Methods Enzymol. 1994;244:350–375. doi: 10.1016/0076-6879(94)44027-1. [DOI] [PubMed] [Google Scholar]
- 28.Kolatka K, Witosinska M, Pierechod M, Konieczny I. Bacterial partitioning proteins affect the subcellular location of broad-host-range plasmid RK2. Microbiology. 2008;154:2847–2856. doi: 10.1099/mic.0.2008/018762-0. [DOI] [PubMed] [Google Scholar]
- 29.Grimminger V, Richter K, Imhof A, Buchner J, Walter S. The prion curing agent guanidinium chloride specifically inhibits ATP hydrolysis by Hsp104. J. Biol. Chem. 2004;279:7378–7383. doi: 10.1074/jbc.M312403200. [DOI] [PubMed] [Google Scholar]
- 30.Toukdarian AE, Helinski DR. TrfA dimers play a role in copy-number control of RK2 replication. Gene. 1998;223:205–211. doi: 10.1016/s0378-1119(98)00370-9. [DOI] [PubMed] [Google Scholar]
- 31.Cereghino JL, Helinski DR, Toukdarian AE. Isolation and characterization of DNA-binding mutants of a plasmid replication initiation protein utilizing an in vivo binding assay. Plasmid. 1994;31:89–99. doi: 10.1006/plas.1994.1009. [DOI] [PubMed] [Google Scholar]
- 32.Fu GK, Markovitz DM. The human LON protease binds to mitochondrial promoters in a single-stranded, site-specific, strand-specific manner. Biochemistry. 1998;37:1905–1909. doi: 10.1021/bi970928c. [DOI] [PubMed] [Google Scholar]
- 33.Fu GK, Smith MJ, Markovitz DM. Bacterial protease Lon is a site-specific DNA-binding protein. J. Biol. Chem. 1997;272:534–538. [PubMed] [Google Scholar]
- 34.Lu B, Liu T, Crosby JA, Thomas-Wohlever J, Lee I, Suzuki CK. The ATP-dependent Lon protease of Mus musculus is a DNA-binding protein that is functionally conserved between yeast and mammals. Gene. 2003;306:45–55. doi: 10.1016/s0378-1119(03)00403-7. [DOI] [PubMed] [Google Scholar]
- 35.Zehnbauer BA, Foley EC, Henderson GW, Markovitz A. Identification and purification of the Lon+ (capR+) gene product, a DNA-binding protein. Proc. Natl Acad. Sci. USA. 1981;78:2043–2047. doi: 10.1073/pnas.78.4.2043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Smith CK, Baker TA, Sauer RT. Lon and Clp family proteases and chaperones share homologous substrate-recognition domains. Proc. Natl Acad. Sci. USA. 1999;96:6678–6682. doi: 10.1073/pnas.96.12.6678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu WF, Zhou Y, Gottesman S. Redundant in vivo proteolytic activities of Escherichia coli Lon and the ClpYQ (HslUV) protease. J. Bacteriol. 1999;181:3681–3687. doi: 10.1128/jb.181.12.3681-3687.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pruteanu M, Baker TA. Controlled degradation by ClpXP protease tunes the levels of the excision repair protein UvrA to the extent of DNA damage. Mol. Microbiol. 2009;71:912–924. doi: 10.1111/j.1365-2958.2008.06574.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frank EG, Ennis DG, Gonzalez M, Levine AS, Woodgate R. Regulation of SOS mutagenesis by proteolysis. Proc. Natl Acad. Sci. USA. 1996;93:10291–10296. doi: 10.1073/pnas.93.19.10291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zylicz M, Liberek K, Wawrzynow A, Georgopoulos C. Formation of the preprimosome protects lambda O from RNA transcription-dependent proteolysis by ClpP/ClpX. Proc. Natl Acad. Sci. USA. 1998;95:15259–15263. doi: 10.1073/pnas.95.26.15259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pruteanu M, Neher SB, Baker TA. Ligand-controlled proteolysis of the Escherichia coli transcriptional regulator ZntR. J. Bacteriol. 2007;189:3017–3025. doi: 10.1128/JB.01531-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Shah IM, Wolf RE., Jr Inhibition of Lon-dependent degradation of the Escherichia coli transcription activator SoxS by interaction with 'soxbox' DNA or RNA polymerase. Mol. Microbiol. 2006;60:199–208. doi: 10.1111/j.1365-2958.2006.05086.x. [DOI] [PubMed] [Google Scholar]
- 43.Abdelhakim AH, Sauer RT, Baker TA. The AAA+ ClpX machine unfolds a keystone subunit to remodel the Mu transpososome. Proc. Natl Acad. Sci. USA. 2010;107:2437–2442. doi: 10.1073/pnas.0910905106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kuroda A, Nomura K, Ohtomo R, Kato J, Ikeda T, Takiguchi N, Ohtake H, Kornberg A. Role of inorganic polyphosphate in promoting ribosomal protein degradation by the Lon protease in E. coli. Science. 2001;293:705–708. doi: 10.1126/science.1061315. [DOI] [PubMed] [Google Scholar]
- 45.Nomura K, Kato J, Takiguchi N, Ohtake H, Kuroda A. Effects of inorganic polyphosphate on the proteolytic and DNA-binding activities of Lon in Escherichia coli. J. Biol. Chem. 2004;279:34406–34410. doi: 10.1074/jbc.M404725200. [DOI] [PubMed] [Google Scholar]
- 46.Lee AY, Hsu CH, Wu SH. Functional domains of Brevibacillus thermoruber lon protease for oligomerization and DNA binding: role of N-terminal and sensor and substrate discrimination domains. J. Biol. Chem. 2004;279:34903–34912. doi: 10.1074/jbc.M403562200. [DOI] [PubMed] [Google Scholar]
- 47.Lin YC, Lee HC, Wang I, Hsu CH, Liao JH, Lee AY, Chen C, Wu SH. DNA-binding specificity of the Lon protease alpha-domain from Brevibacillus thermoruber WR-249. Biochem. Biophys. Res. Commun. 2009;388:62–66. doi: 10.1016/j.bbrc.2009.07.118. [DOI] [PubMed] [Google Scholar]
- 48.Guo F, Maurizi MR, Esser L, Xia D. Crystal structure of ClpA, an Hsp100 chaperone and regulator of ClpAP protease. J. Biol. Chem. 2002;277:46743–46752. doi: 10.1074/jbc.M207796200. [DOI] [PubMed] [Google Scholar]
- 49.Charette MF, Henderson GW, Doane LL, Markovitz A. DNA-stimulated ATPase activity on the lon (CapR) protein. J. Bacteriol. 1984;158:195–201. doi: 10.1128/jb.158.1.195-201.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chung CH, Goldberg AL. DNA stimulates ATP-dependent proteolysis and protein-dependent ATPase activity of protease La from Escherichia coli. Proc. Natl Acad. Sci. USA. 1982;79:795–799. doi: 10.1073/pnas.79.3.795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sonezaki S, Okita K, Oba T, Ishii Y, Kondo A, Kato Y. Protein substrates and heat shock reduce the DNA-binding ability of Escherichia coli Lon protease. Appl. Microbiol. Biotechnol. 1995;44:484–488. doi: 10.1007/BF00169948. [DOI] [PubMed] [Google Scholar]
- 52.Kuroda A. A polyphosphate-lon protease complex in the adaptation of Escherichia coli to amino acid starvation. Biosci. Biotechnol. Biochem. 2006;70:325–331. doi: 10.1271/bbb.70.325. [DOI] [PubMed] [Google Scholar]
- 53.Simmons LA, Grossman AD, Walker GC. Clp and Lon proteases occupy distinct subcellular positions in Bacillus subtilis. J. Bacteriol. 2008;190:6758–6768. doi: 10.1128/JB.00590-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.