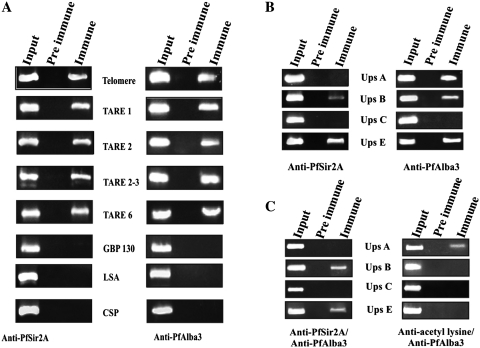
Figure 7.
ChIP and ChIP ReChIP analysis (A) ChIP analysis performed from ring stage parasites using antibodies against PfSir2A (left panel) and PfAlba3 (right panel). Immunoprecipitated DNA was analyzed by PCR amplification specific to telomeric and subtelomeric sequences (TARE1, TARE2, TARE2-3 and TARE6), GBP130, LSA and CSP 5′-UTR region. Input DNA material was used as a positive control, whereas material immunoprecipitated by the pre-immune sera or IgG used as control. PfAlba3 and PfSir2A are assembled at the telomeric and subtelomeric regions of chromatin, as shown by ChIP analysis. (B) PfAlba3 is also associated to the 5′-UTR of var genes. ChIP analysis of ring stage parasites was performed using antibodies against PfSir2A (left panel) and PfAlba3 (right panel). (C) PfAlba3 and PfSir2A are assembled at same promoter region and the association between PfSir2A and PfAlba3 causes its deacetylation, as shown by ChIP ReChIP analysis. Soluble chromatin was prepared from isolated parasites and divided into two chromatin aliquots, which were immunoprecipitated with antibodies to either anti-PfSir2A/PfAlba3 antibody or anti-acetyl lysine/PfAlba3 antibody, respectively. Controls for the ChIP ReChIP fractions include the input chromatin and pre-immune serum. Input corresponds to DNA prepared from fragmented chromatin prior to immunoprecipitation. Each experiment was performed in triplicate and the data presented were a representation of one out of three experiments showing similar result.