Abstract
Transcriptional regulation plays a critical role in the life cycle of Mycobacterium smegmatis and its related species, M. tuberculosis, the causative microbe for tuberculosis. However, the key transcriptional factors involved in broad regulation of diverse genes remain to be characterized in mycobacteria. In the present study, a TetR-like family transcriptional factor, Ms6564, was characterized in M. smegmatis as a master regulator. A conserved 19 bp-palindromic motif was identified for Ms6564 binding using DNaseI footprinting and EMSA. A total of 339 potential target genes for Ms6564 were further characterized by searching the M. smegmatis genome based on the sequence motif. Notably, Ms6564 bound with the promoters of 37 cell cycle and DNA damage/repair genes and regulated positively their expressions. The Ms6564-overexpressed recombinant strain yielded 5-fold lower mutation rates and mutation frequencies, whereas deletion of Ms6564 resulted in ∼5-fold higher mutation rates for the mutant strain compared with the wild-type strain. These findings suggested that Ms6564 may function as a global regulator and might be a sensor necessary for activation of DNA damage/repair genes.
INTRODUCTION
Transcriptional regulation plays an important role in the life cycle of Mycobacterium tuberculosis (1,2), the causative microbe for tuberculosis (TB), which results in the death of ∼2 million people globally each year (3). A unique DNA damage/repair mechanism has been proposed in M. tuberculosis (4). However, the regulations and consequence of these genes remain largely unclear. Mycobacterium smegmatis is a fast-growing non-pathogenic mycobacterium widely used as a model organism to study the biology of other virulent and extremely slow growing species like M. tuberculosis (5). In particular, the genome of M. smegmatis encodes more than 500 regulatory factors (GenBank accession number CP000480), which are strikingly more than the ∼180 encoded by M. tuberculosis (1).
Generally, bacteria respond to DNA damage through an increase in the expression of a number of genes, resulting in a greater rate of survival. This response is regulated by the homologs of the Escherichia coli repressor protein LexA in many species (6). At least two mechanisms for DNA damage induction exist in M. tuberculosis (7); a LexA-regulated system dependent on RecA and a RecA/LexA-independent mechanism for DNA damage induction, which has yet to be characterized clearly (7). A few other genes have been reported to be upregulated in E. coli following DNA damage independent of LexA (8) or RecA (9). Interestingly, a global analysis of gene expression following DNA damage in both the wild-type strain and recA deletion mutant of M. tuberculosis demonstrated that the majority of inducible DNA repair genes in M. tuberculosis were induced independently of RecA (10). However, the target genes controlled by the majority of the transcription factors and the functional roles of these regulations in vivo remain largely unknown.
TetR is a large family of transcriptional regulators. Its prototype is TetR from the Tn10 transposon of E. coli, which functions to regulate the expression of a tetracycline efflux pump in Gram-negative bacteria (11). These proteins often serve as repressors and are widely distributed among bacteria, regulating a number of diverse processes (12). For example, Staphylococcus aureus QacR regulates the expression of a multidrug transporter (13). Mycobacterium tuberculosis EthR regulates the expression of a monooxygenase gene that catalyzes the activation of ethionamide, an antibiotic used in TB treatment (14,15). KstR, a highly conserved transcriptional repressor, in M. smegmatis and M. tuberculosis which also belongs to the TetR family, directly controls the expression of 83 genes in M. smegmatis and 74 genes in M. tuberculosis (16). SczA is one of the few examples of regulators from the TetR family that function as a transcriptional activator (17).
In the present study, a new TetR family transcriptional regulator, Ms6564, was examined in M. smegmatis. Evidence was provided to show that Ms6564 is a candidate for the broad regulation of gene expression including cell cycle and RecA-dependent and RecA-independent DNA damage/repair genes. In particular, Ms6564 was demonstrated to function as a master activator and a negative regulator of gene mutation rates.
MATERIALS AND METHODS
Strains, enzymes, plasmids and reagents
E. coli BL21 cells and pET28a were purchased from Novagen and were used to express mycobacterial proteins. pBT, pTRG vectors and E. coli XR host strains were purchased from Stratagene. Restriction enzymes, T4 ligase, modification enzymes, Pyrobest DNA polymerase, dNTPs and all antibiotics were obtained from TaKaRa Biotech. The reagents for one-hybrid assay were purchased from Stratagene. Polymerase Chain Reaction (PCR) primers were synthesized by Invitrogen (Supplementary Table S1) and Ni-NTA (Ni2+-nitrilotriacetate) agarose was obtained from Qiagen.
Cloning of M. smegmatis transcription factors and regulatory sequences of the target genes and bacterial one-hybrid assays
About 505 transcription factors were predicted from the genome of M. smegmatis mc2 155 National Center of Biotechnology Information. All of these probable genes were amplified using their respective primers and were cloned into the pTRG vector (Stratagene). A subgenomic library for M. smegmatis mc2 155 transcription factors was produced by mixing these recombinant plasmids. The promoters of the M. smegmatis mc2 155 genes were also amplified using their primers (Supplementary Table S1) and were cloned into pBXcmT vector (2). E. coli XL1-Blue MRF′ Kan strain (Stratagene) was used for the routine propagation of all pBXcmT and pTRG recombinant plasmids. BacterioMatch I One-Hybrid System (Stratagene) was utilized to detect DNA–protein interactions between pBXcmT and pTRG plasmids as described previously (2). The recombinant plasmid pBXcmT was used to screen the library for M. smegmatis mc2 155 transcription factors. Positive growth co-transformants were selected on a selective screening medium plate containing 20 mM 3-AT, 16 μg/ml streptomycin, 15 μg/ml tetracycline, 34 μg/ml chloramphenicol and 50 μg/ml kanamycin. The plates were incubated at 30°C for 3–4 days. A co-transformant containing pBX-R2031/pTRG-R3133 plasmids (2) was served as positive control and a co-transformant containing empty vector pBX and pTRG was also served as negative control.
Expression and purification of recombinant proteins
Mycobacterium smegmatis mc2 155 genes were amplified by PCR primers from genomic DNA (Supplementary Table S1). The corresponding genes were cloned into pET28a to produce recombinant vectors. Transformed with the recombinant plasmid, E. coli BL21 cells were grown in a 200 ml LB medium up to an OD600 of 0.6. Protein expression was induced by the addition of 0.5 mM isopropyl β-D-1-thiogalactopyranoside (IPTG). The harvested cells were resuspended and sonicated in binding buffer (100 mM Tris–HCl pH 8.0, 500 mM NaCl and 10 mM imidazole) for his-tagged proteins. The lysate was centrifuged at 10 000g for 30 min, and the cleared supernatant was loaded on the affinity column. The column-bound protein was washed with a wash buffer (100 mM Tris–HCl pH 8.0, 500 mM NaCl and 40 mM imidazole) for his-tagged proteins. The protein was then eluted using an elution buffer (100 mM Tris–HCl pH 8.0, 500 mM NaCl and 250 mM imidazole) for his-tagged proteins. The elution was dialyzed overnight and stored at −80°C. Protein concentration was detected by Coomassie Brilliant Blue assay.
DNA substrate preparation and electrophoretic mobility shift assay
The DNA fragments for the DNA-binding activity assays were amplified by PCR from M. smegmatis mc2 155 genomic DNA or directly synthesized by Invitrogen (Supplementary Table S2). The amplified products were purified with BioFlux PCR DNA Purification kit (BioFlux) labeled with T4 polynucleotide kinase (Takara) and [γ-32P] Adenosine Triphosphate (ATP) following the manufacturer's instructions. The mixture was treated at 65°C for 7 min to inactivate the protein kinase in the reactions. The labeled DNA substrates were then stored at −20°C until use. The synthesized oligonucleotide was radioactively labeled with T4 polynucleotide kinase (Takara) and [γ-32P] ATP. The labeled oligonucleotide was purified as described previously (18). The 1.2-fold unlabeled reverse oligonucleotide was added and incubated at 95°C for 10 min to allow complete annealing. The DNA substrates were stored at −20°C until use. Labeled DNA fragments were incubated at 25°C for 30 min or 1 h with various amounts of proteins in a total volume of 20 μl electrophoretic mobility shift assay (EMSA) buffer consisting of 50 mM Tris–HCl pH 7.5, 10 mM MgCl2, 1 mM DTT and 50 mM NaCl. The mixtures were then directly subjected to 5% native Polyacrylamide Gel Electrophoresis containing 0.5×Tris–borate–EDTA buffer. Electrophoresis was performed at 150 V at 25°C. Images were acquired by Typhoon Scanner (GE Healthcare).
DNase I footprinting assays
The 189 bp promoter regions Ms6564p4-1 (coding strand) and Ms6564p4-2 (non-coding strand) (Supplementary Table S2) were amplified by PCR using their primers labeled with Fluorescein Isothiocyanate (Supplementary Table S1). The amplified products were purified with BioFlux PCR DNA Purification kit (BioFlux) and then subjected to the same binding reaction as in EMSA. DNaseI footprinting was performed as described previously (19). The ladders were produced using the Sanger dideoxy method and Ms6564p4f1 and Ms6564p4r2 primers (Supplementary Table S1).
Construction of the Ms6564 deletion mutant of M. smegmatis mc2155 and Southern blot analysis
Knockout of the Ms6564 gene from M. smegmatis mc2155 (20) was performed as described previously (21). A pMind (22) derived suicide plasmid carrying a hygromycin resistance gene was constructed and a sacB gene was inserted to confer sensitivity to sucrose as a negative selection marker. The recombinant plasmid pMindMs6564 was electrophorated into M. smegmatis mc2155 and selected on 7H10 medium containing 100 µg/ml hygromycin and 4% sucrose. Genomic DNA from allelic-exchange mutants in which the Ms6564 gene had been deleted was identified by restriction digestion and confirmed by PCR analysis using the primers on each side of Ms6564 and the hygromycin gene.
The deleted Ms6564 gene was identified by Southern blot analysis. Approximately 10 µg genomic DNA was digested overnight with an excess of NarI, and the fragments were separated by electrophoresis through 0.8% agarose gels. Southern blotting was carried out in 10× Saline Sodium Citrate (SSC) using Hybond-N+ nylon membranes (Amersham). The probe consisted of a 392 bp fragment of the upstream region of the Ms6564 gene amplified using a pair of its primers (Supplementary Table S1). The Prime a Gene labeling system (Amersham) and 5 µCi digolan were used to label the probe. Prehybridization and hybridization were carried out at 65°C using 5×SSC, 5×Denhardt's solution and 0.5% Sodium Dodecyl Sulfate (SDS). Serial 15 min washes were performed at 65°C as follows: two washes with 2×SSC and 0.1% SDS and two washes with 1×SSC and 1% SDS. The filter was developed and photographed.
Quantitative real-time PCR
Isolation of mRNA and cDNA from Msm/pMV261 and Msm/pMV261-Ms6564 (Msm/WT and Msm/Ms6564::hyg) strains was performed as described previously (23). For real-time PCR analysis, gene-specific primers (Supplementary Table S3) were used, and first-strand cDNAs were synthesized using SuperScript II reverse transcriptase (Invitrogen) according to the manufacturer's instructions. Each PCR reaction (20 μl) contained 10 μl of 2×SYBR Green Master Mix Reagent (Applied Biosystems), 1.0 μl of cDNA samples and 200 nM gene-specific primers. The reactions were performed in Bio-Rad IQ5 RT-PCR machine. The thermocycling conditions were 95°C for 5 min and 40 cycles at 95°C for 30 s, 60°C for 30 s and 72°C for 30 s. Amplification specificity was assessed using melting curve analysis. Different gene expressions were normalized to the levels of 16S rRNA gene transcripts (24). The degrees of expression change were calculated using the 2−ΔΔCt method (25).
Quantitative chromatin immunoprecipitation
Chromatin immunoprecipitation (ChIP) was performed as described previously (26). Mycobacterium smegmatis mc2 155 cells were grown in a 100 ml 7H9 medium up to an OD600 of 1.0, fixed with 1% formaldehyde for 20 min and stopped with 0.125 M glycine for 5 min. Crosslinked cells were harvested and resuspended in 1 ml Tris-Buffered Saline supplemental with Tween-20 and Triton-X 100 (20 mM Tris-HCl [pH 7.5], 150 mM NaCl, 0.1% Tween 20, 0.1% Triton X-100). The sample was sonicated on ice and the average DNA fragment size was determined to be ∼0.5 kb. A 100 µl sample of the extract was saved as the input fraction, whereas the remaining 900 µl was incubated with 10 µl of antibodies against Ms6564 or preimmune serum under rotation for 3 h at 4°C. The complexes were immunoprecipitated with 20 µl 50% protein A agarose for 1 h under rotation at 4°C. The immunocomplex was recovered by centrifugation and resuspended in 100 µl TE (20 mM Tris–HCl pH 7.8, 10 mM EDTA, 0.5% SDS). Crosslinking was reversed for 6 h at 65°C. The DNA samples of the input and ChIP were purified, resuspended in 50 µl TE and analyzed by PCR with Platinum Taq (Invitrogen). Each experiment was performed in duplicate and repeated twice. The amplification protocol included one denaturation step of 5 min at 95°C, then 32 cycles of 1 min at 95°C, 1 min at 60°C and 1 min at 72°C.
Analysis of β-galactosidase activity
β-galactosidase activity experiments were performed in M. smegmatis by creating operon-lacZ fusions based on the expression vector of pMV261 (27). Promoter sequences were first cloned into pMV261 backbone by XbaI/EcoRI and then the reporter gene lacZ was cloned by HindIII/NheI. The reporter plasmids were transformed into mutant ΔMs6564 strain to obtain the corresponding recombinant reporter strains ΔY0, ΔY1, ΔY2, ΔY3, ΔY4, ΔY5 and ΔY6. They were transformed into wild-type M. smegmatis to obtain the corresponding reporter strains Y0, Y1, Y2, Y3, Y4, Y5 and Y6 (Supplementary Table S4). All strains were grown in 7H9-Tw-glycerol-Kan medium at 37°C for 48 h (23). Some cell suspension was then incubated into 7H9-Tw-glycerol-Kan liquid medium and grown at 37°C to an OD600 of 0.5–0.8. β-Galactosidase measurements were performed as described previously (28). Another cell suspension was plated on 7H10- glycerol-Kan-X-gal solid medium and grown at 37°C for imaging.
Estimation of mutation frequencies and rates
The mutation frequency of the streptomycin-resistant gene in both the wild-type strain and mutant strains was examined as reported previously (29). Briefly, single colonies from M. smegmatis strains were grown in Middlebrook 7H9 medium (Difco) supplemented with 10% (v/v) albumin-dextrose-catalase (Merck), 0.2% glycerol and 0.1% Tween-80 to an optical density at 600 nm of 1.2–1.5. Then 1 ml of the cultures were plated in triplicate onto a 7H10 solid medium containing Str (50 μg/ml) to monitor spontaneous mutation frequencies. The remaining cultures were diluted to 10−6, and 200 μl of the dilution was plated in triplicate onto a 7H10 antibiotic-free solid medium for Colony-forming unit (CFU) determination. Mutation frequencies were calculated as reported previously (29).
The rates of the spontaneous mutation of M. smegmatis strains to streptomycin (Str) resistance were determined by Luria–Delbrück fluctuation analysis (30) using the method described by Machowski et al. (31) with slight modification. Single colonies from M. smegmatis wild-type strain, Ms6564 deletion strain and Ms6564 deletion strain complemented with a Ms6564 expression plasmid pMindD6564 were grown in Middlebrook 7H9 medium (Difco) supplemented with 10% (v/v) albumin-dextrose-catalase (Merck), 0.2% glycerol and 0.1% Tween-80 to an optical density at 600 nm of 1.2–1.5. To stabilize pMindD6564 in the complemented mutant strain, additional kanamycin (50 μg/ml) was placed in the medium. For the selection of mutant clones, the entire contents of a culture tube were plated on Str-supplemented medium following the removal of a 100 μl aliquot for CFU determination. The final number of cells in the culture (Nt) and the observed number of mutant in the same culture (r) were determined by plating with and without streptomycin, respectively. Mutation rates were calculated by the method of the median (32) using the following formula: mutation rate = m/Nt, where m was calculated from r via the Lea–Coulson equation: r/m − ln(m) − 1.24 = 0 using the web tool Fluctuation AnaLysis CalculatOR (33).
RESULTS
Ms6564 interacts with the promoters of RecA-dependent and RecA-independent DNA repair genes and its own promoter region
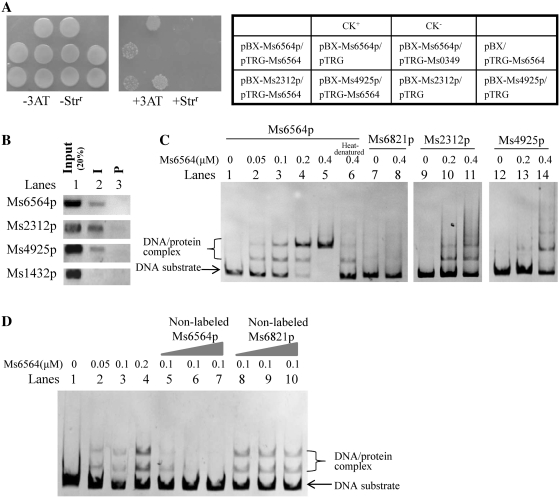
We used a bacterial one-hybrid system (2), which detected protein–DNA interactions based on transcriptional activation of reporter genes of HIS3 and aadA, to search for the potential common transcriptional factors involved in the broad regulation of the expressions of both RecA-dependent and RecA-independent DNA repair genes in M. smegmatis. The promoter region of Ms2312, a reported RecA-dependent DNA repair gene (10) or Ms4925, a RecA-independent gene was cloned into the upstream of HIS3–aadA in the reporter vector pBXcmT (2). The library of predicted transcriptional regulators from M. smegmatis was screened using these two promoters, Ms2312p and Ms4925p, as a bait sequence. In a bacterial one-hybrid assay, a putative TetR-like transcriptional factor, Ms6564, interacted with both promoters in Figure 1A. This result was evident in the co-transformants with pTRG-Ms6564/pBX-Ms2312p and pTRG-Ms6564/pBX-Ms4925p that grew very well in the screening medium. The positive control, composed of co-transformants with pTRG-Rv3133c/pBX-Rv2031p (2), also grew well in the medium (Figure 1A). The binding of Ms6564 with its own promoter was likewise examined because most members of the TetR family presented an auto-regulation mechanism. The co-transformants with pTRG-Ms6564/pBX-Ms6564p grew very well in the screening medium as shown in Figure 1A. By contrast, no growth was observed for their self-activated controls. Therefore, Ms6564 can bind to the promoters of RecA-dependent and RecA-independent DNA repair genes and it can also bind with its own promoter.
Figure 1.
Interaction of Ms6564 with the promoter regions of RecA-dependent and RecA-independent DNA repair genes and its own promoter region. (A) Bacterial one-hybrid assays. The promoter of the Ms2312, Ms4925 and Ms6564 genes was cloned into pBXcmT, and Ms6564 was cloned into pTRG vectors. A pair of pBXcmT/pTRG plasmids was co-transformed into the reporter strain and then its growth was tested together with the self-activation controls on a selective medium containing 3-AT, Kanr, Strr and Chlr as described in ‘Materials and Methods’ section. An outline of the plates is shown in the right panel. Each unit represents the corresponding co-transformant in the plates. (B) ChIP assays. ChIP using preimmune (P) or immune sera (I) raised against Ms6564. Exponentially growing M. smegmatis cells were fixed with 1% formaldehyde. Crosslinked cells were resuspended and sonicated on ice. A 100 µl sample of the extract was saved as the input fraction, with the remaining 900 µl incubated with 10 µl of antibodies against Ms6564 at 4°C. The complexes were immunoprecipitated with 20 µl 50% protein A-agarose. The immunocomplex was recovered by centrifugation and resuspended in 100 µl TE. Crosslinking was reversed for 6 h at 65°C. The DNA samples of the input and ChIP were purified and resuspended in 50 µl TE. Then the DNA recovered from the immunoprecipitates was amplified with primers specific for DNA repair genes or to an unrelated mycobacterial promoter of Ms1432 used as a negative control. (C) EMSA assays. 32P-labeled Ms6564p (lanes 1–5), Ms2312p (lanes 9–11), Ms4925p (lanes 12–14) or Ms6821p (a non-specific DNA, lanes 7 and 8) DNA substrates were co-incubated with various amounts of Ms6564 protein. The heat-denatured Ms6564 was used as negative control (lane 6). The free DNA substrate and DNA–protein complex are indicated. (D) EMSA assays for the specific binding of Ms6564 with the DNA substrate. Unlabeled cold Ms6564 or unspecific Ms6821 promoter DNA substrates were used to compete with the [γ-32P] ATP labeled Ms6564 promoter DNA. Cold Ms6564 promoter DNA, but not Ms6821 promoter DNA, could competitively inhibit the binding of Ms6564 to the labeled Ms6564 promoter DNA substrate.
Ms6564 specifically binds to the target promoters both in vivo and in vitro
ChIP assay was subsequently conducted to examine the binding of Ms6564 to Ms2312p, Ms4925p and Ms6564p in vivo. As shown in Figure 1B, Ms6564 can be crosslinked to Ms2312p, Ms4925p and Ms6564p. These promoter DNAs can be particularly recovered by immunoprecipitation through the specific Ms6564 antiserum (Figure 1B, lane 2). By contrast, the preimmune serum failed to precipitate significant amounts of DNA (Figure 1B, lane 3). Ms1432p, the promoter of an unrelated gene used as negative control, cannot be recovered by the Ms6564 antiserum. Further EMSA assays confirmed the binding of the purified Ms6564 protein to these target promoter DNAs in vitro. As shown in Figure 1C, when 3 nM Ms6564 promoter DNA substrates were co-incubated with increasing amounts of Ms6564 (0, 0.05, 0.1, 0.2 and 0.4 µM), clear shifted bands were observed (Figure 1C, lanes 2–5). By contrast, the heat-denatured Ms6564 protein lost most of its binding activities (Figure 1C, lane 6). Ms6564 cannot bind with an unrelated Ms6821p promoter DNA (Figure 1C, lanes 7 and 8). Therefore, Ms6564 can bind with its promoter DNA. It can also bind with Ms2312p (Figure 1C, lanes 9–11) and Ms4925p (Figure 1C, lanes 12–14) forming a clear protein–DNA complex on the gel. A competition assay confirmed the specificity of Ms6564 binding with its promoter DNA. Unlabeled cold Ms6564 or unspecific Ms6821 promoter DNA substrates were used to compete with the labeled Ms6564 promoter DNA. As shown in Figure 1D, cold Ms6564 promoter DNA, but not Ms6821 promoter DNA, could competitively inhibit the binding of Ms6564 to the labeled Ms6564 promoter DNA substrate.
The above findings strongly suggested that Ms6564 can bind with the promoters of both RecA-dependent and RecA-independent DNA repair genes, as well as its own promoter region.
Ms6564 binds with DNA fragments containing a palindrome sequence motif
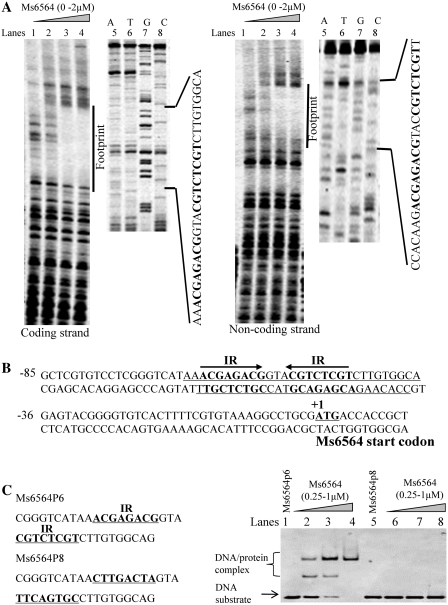
A series of truncated DNA substrates within the promoter region of Ms6564, designated as p1–p7 (Supplementary Figure S1A), was produced to characterize the DNA-binding motif for Ms6564 protein. After two cycles of EMSA assays, the binding region was mapped further into Ms6564 p6 as evidenced by an obvious DNA-binding activity on the 39 bp-length substrate p6, but not on p5 or p7 (Supplementary Figure S1B).
The binding motif for the recognition of Ms6564 was characterized by further DNaseI footprinting assays. As shown in Figure 2A, when increasing amounts of Ms6564 protein (0–2 µM) were co-incubated with DNaseI, the region around AACGAGACGGTACGTCTCGT was obviously protected on the coding strand. This result indicates that the DNA fragment contained a potential binding motif for Ms6564. Similarly, the region around CCACAAGACGAGACGT ACCGTCTCGTT was protected when the non-coding strand DNA was used as substrate (Figure 2A, right panel). The protected DNA region was extended from position −66 to −37 in the coding strand and from position −65 to −39 in the non-coding strand (Figures 2B and C). A palindromic motif formed by two inverted repeats (IR, 5′-ACGAGACG-3′) separated from each other by three nucleotides (Figures 2B and C) was found from an analysis of this protected sequence. Further EMSA assays were conducted to confirm the significance of the motif for the specific recognition by Ms6564. As shown in Figure 2C (right panel, lanes 5–8), Ms6564 lost the capability to bind with the Ms6564-p8 in which two inverted repeats were replaced by the random sequences CTTGACTA and TTCAGTGC. Therefore, the putative binding sites for Ms6564 contained a specific palindromic sequence motif.
Figure 2.
DNA-binding motif assays for Ms6564. (A) DNaseI footprinting experiments. The assay of the protection of Ms6564 promoter DNA was performed against DNaseI digestion by increasing the amount of Ms6564 (lanes 1–4). The ladders are shown and the corresponding nucleotide sequence is listed (lanes 5–8). The protected regions on the coding strand (left panel) and non-coding strand (right panel) are indicated by a black bar. (B) Sequence and structural characteristics of the protected Ms6564 promoter region. The regions protected by Ms6564 are shown with underlines and the box highlights the 19 bp sequences containing the invert repeat (IR) with 3 bp separations. The translation start codon of Ms6564 is indicated in bold. (C) EMSA assays for the DNA-binding activity of Ms6564 on the DNA substrates with (lanes 1–4) or without the IR sequence (lanes 5–8). Either DNA substrate was co-incubated with 0.25–1 µM Ms6564 protein.
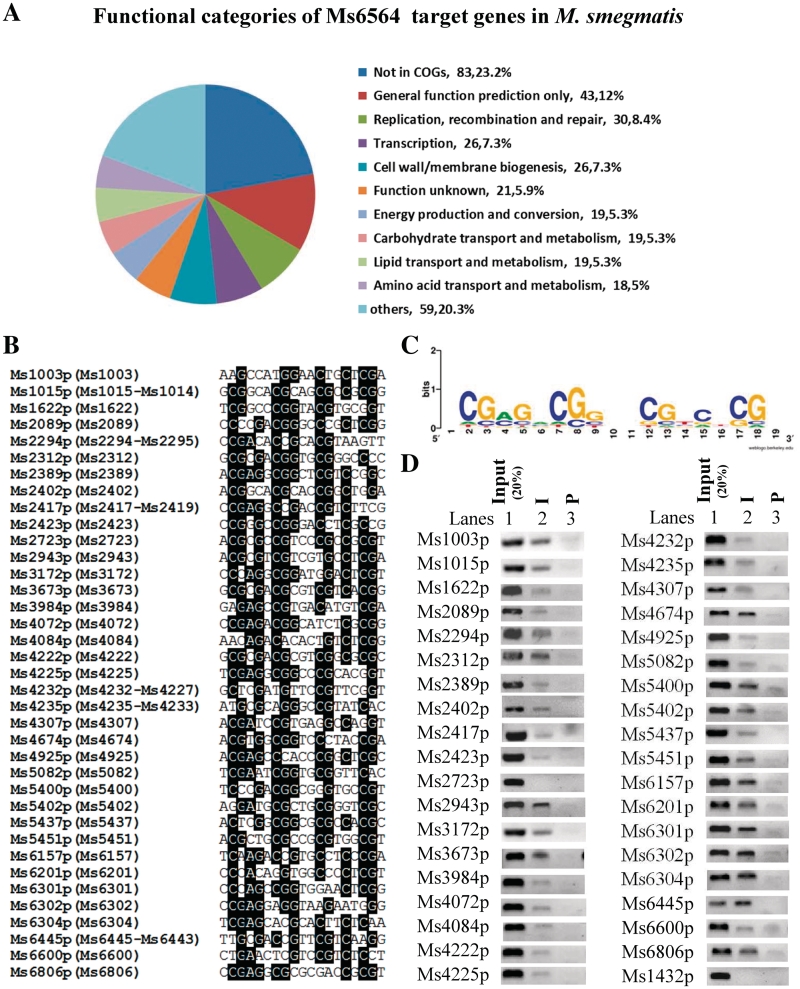
Ms6564 binds with the promoters of many DNA damage/repair and cell cycle genes
The intergenic regions of the M. smegmatis genome were searched based on the sequence motif. A total of 339 potential target genes were characterized (Supplementary Table S5 and Supplementary Figure S2). We further analyzed the classification and percentage of these target genes in the context of Cluster of Orthologous Groups of proteins (COG) categories. As shown in Figure 3A and B, among several defined functional categories, notably, it included 37 promoters which regulated expression of cell cycle and DNA damage/repair genes. Using the WebLogo tool (34), a logo assay was conducted to search for a more general conserved motif for Ms6564 binding. An inverted repeat sequence was characterized within the motif as shown in Figure 3C. Further in vivo ChIP assay established that 29 of all 30 target promoters of DNA damage/repair genes and seven cell cycle genes can be specifically recovered by immunoprecipitation through specific Ms6564 antiserum (Figure 3D). Only one promoter, Ms2723p, was not validated successfully. A negative control, Ms1432p, cannot be recovered by the Ms6564 antiserum. Therefore, these results proved that Ms6564 can regulate a large number of target genes.
Figure 3.
Target promoter sequence analysis of DNA damage and repair genes and in vivo DNA-binding activity of Ms6564 assays. (A) Functional categories of Ms6564 target genes in M. smegmatis. The classification and percentage of the target genes were analyzed in the context of COG categories. (B) The IR-containing motif (ACGAGACGGTA CGTCTCGT) was used to search the intergenic regions between M. smegmatis Open Reading Frames (ORFs). The identified promoters of the DNA damage and repair genes containing the motif are blasted and listed. The conserved sequence is highlighted by black backgrounds. The included genes in the same operon were presented in the following brackets. (C) Logo assays for the protected region. The logos were generated by MEME software suite (34). (D) ChIP assays for the association of Ms6564 with DNA damage and repair target genes. ChIP using preimmune (P) or immune sera (I) rose against Ms6564. The DNA samples of the input and ChIP were purified and resuspended in 50 µl TE. DNA recovered from the immunoprecipitates was amplified with primers specific for DNA repair genes or to an unrelated mycobacterial promoter of Ms1432.
Ms6564 positively regulates the expression of DNA damage/repair and cell cycle genes
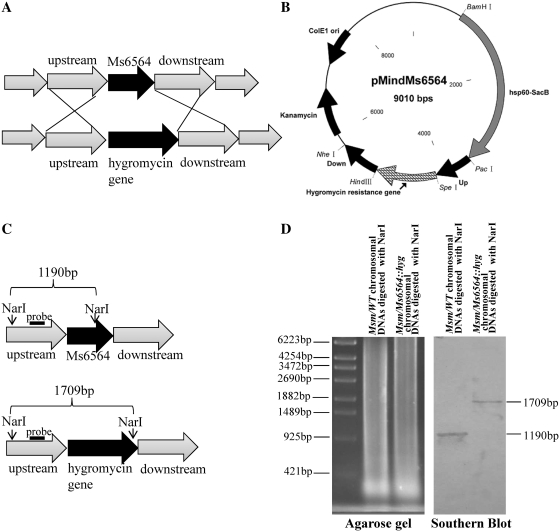
An Ms6564-deleted mutant M. smegmatis strain was produced by gene replacement strategy (Figures 4A and B) to examine further the regulation of Ms6564 on the target genes. A knockout plasmid containing the Up and Down regions of the Ms6564 gene and the selective hygromycin resistance gene (Hgr) was constructed and transformed into M. smegmatis. A ΔMs6564 strain in which the Ms6564 gene was deleted was successfully produced using this method (Figure 4C). Southern blot assay was then conducted to confirm the deletion of Ms6564 in the ΔMs6564 strain. As shown in Figure 4D, a signal band of ∼1.7 kb was detected (Figure 4D, right panel) using a 317 bp probe from the NarI-digested genomic DNA of the mutant M. smegmatis strain. By contrast, a signal band of only ∼1.2 kb was seen in the wild-type strain (Figure 4D, right panel). This finding is consistent with the band sizes expected upon replacement of the Ms6564 gene with the Hygromycinr gene, indicating that the Ms6564 gene was successfully deleted in the mutant strain.
Figure 4.
Construction of the Ms6564 knockout strain of M. smegmatis and Southern blot assays. (A) Schematic representation of the recombination strategy for the removal of Ms6564 from the genome of M. smegmatis. (B) A map of the recombinant vector pMindMs6564 containing upstream and downstream sequences of Ms6564, and the gene that confers resistance against hygromycin. (C) Schematic representation of the DNA fragments of the Msm/WT strain and Msm/ΔMs6564 knockout strain treated with restriction enzyme NarI. The probe is indicated with a black bar. (D) Southern blot assays. A 392 bp probe corresponding to the sequences of the Ms6564 upstream genomic fragment of M. smegmatis was obtained by PCR and labeled with digoxigenin dUTP (Boehringer Mannheim, Inc., Germany). The probe was used to detect the size change of the NarI-digested genomic fragment of M. smegmatis before and after recombination.
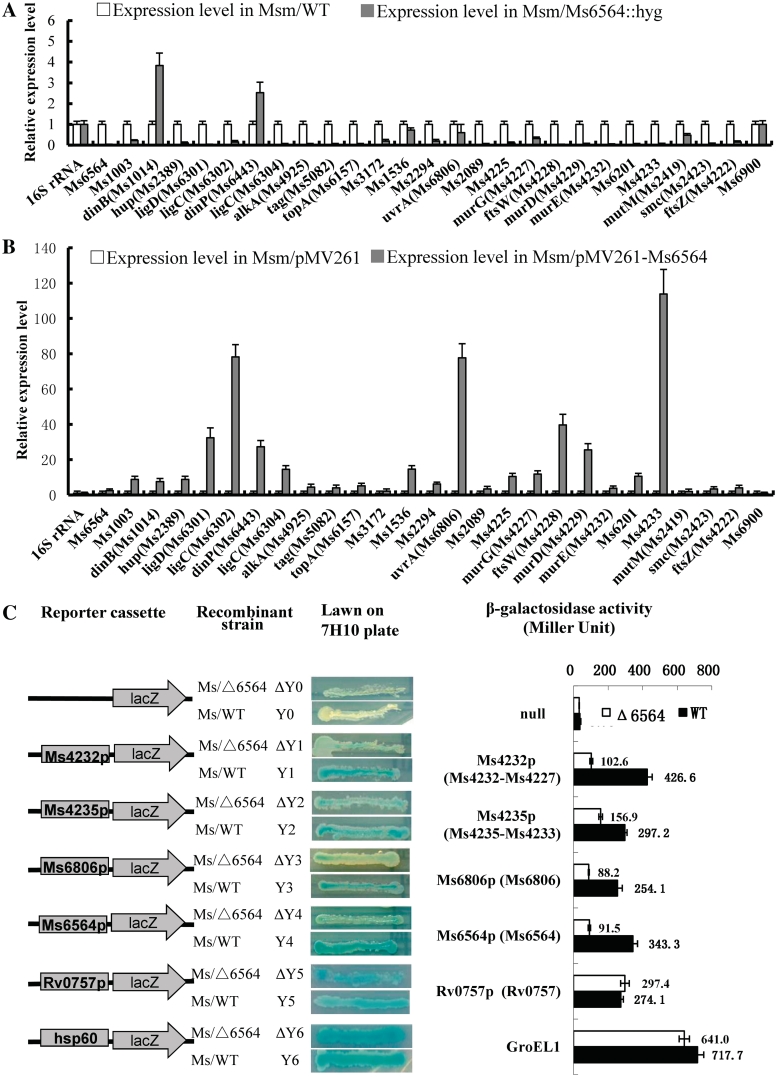
A comparison of the expressions of some cell cycle and DNA damage/repair genes in both wild-type and Ms6564-deleted mutant mycobacterial strains was conducted using quantitative real-time (qRT)-PCR assays. As shown in Figure 5A, compared with the expression in the wild-type strain, those of most of the tested genes were significantly downregulated (P-value < 0.05) in the ΔMs6564 M. smegmatis strains, with the exceptions of dinB and dinP. By contrast, the expression of the negative control gene, Ms6900, had not significantly change. This finding suggested that although two genes were upregulated unexpectedly, Ms6564 can function as a positive regulator for most target genes in M. smegmatis (Figure 5A). Further overexpression assay was conducted to examine the regulatory function of Ms6564. As shown in Figure 5B, the expressions of all tested genes were significantly upregulated (P-value < 0.05) when Ms6564 was overexpressed (∼5-fold) through a pMV261-derived recombinant plasmid in M. smegmatis strains compared with the wild-type strain. This finding is consistent with the above assay in the ΔMs6564 M. smegmatis strains. Relative gene expression levels in response to DNA damage were also measured by qRT-PCR before and after induction by 5 mM H2O2 for 3 h in the ΔMs6564 or wild-type M. smegmatis strains (Supplementary Figure S3). Most of these cell cycle and DNA damage/repair target genes were found to be DNA damage inducible.
Figure 5.
Expression assays of DNA damage and repair genes in wild-type and Ms6564-deleted mutant strains. qRT-PCR assay for the relative expression levels of DNA damage and repair genes in ΔMs6564 M. smegmatis strains (A) and in Ms6564-overexpressed strains (B). The mycobacterial cDNA was amplified as described in Materials and Methods’ section. The relative expression levels of the genes were normalized using 16S rRNA gene as an invariant transcript, and an unrelated promoter gene Ms6900 was used as negative control. Data were analyzed using the 2ΔΔCt method as described previously (25). As a positive control, total DNA of each strain was used as template for PCR amplification. The cDNA of the mutant strains and the recombinant strain containing an empty pMV261 vector was used as template in the negative controls. The P-values of the relative expression data were calculated by unpaired two-tailed Student's t-test using GraphPad Prism 5. (C) The effect of Ms6564 on the gene expression of representative DNA repair genes was assayed by constructing a series of lacZ alone or promoter-lacZ co-expression plasmids. The included genes in the same operon were presented in the following brackets. The activity of β-galactosidase was further examined and presented as Miller units (right panel). Left column: schematic representation of each clone used to generate strains ΔY0–Y6. Null promoter-lacZ, Rv0757p-lacZ and hsp60-lacZ were used as controls. Middle column: exponentially growing M. smegmatis cultures of ΔY0–Y6 were scribed onto 7H10 plates containing 30 µg/ml kanr and 50 µg/ml X-gal. The plates were incubated subsequently for 3–4 days. Right column: β-galactosidase activity was expressed as Miller units. The values presented were the averages of three independent experiments. For statistical analysis, two-way analysis of variance with Bonferroni multiple comparison tests were performed using a P-value of ≤0.05.
A series of promoter-lacZ reporter plasmids was constructed using β-galactosidase as reporter gene in M. smegmatis to confirm further the positive regulation of Ms6564 on the target gene expressions. As shown in Figure 5C, the strong promoter hsp60 strikingly promoted the expression of lacZ in both wild-type and ΔMs6564 M. smegmatis strains compared with the non-promoter lacZ plasmid. The strain appeared deep blue and had high β-galactosidase (∼700 Miller units), indicating that the report system worked well (Figure 5C, bottom of the panel). Six promoters including Ms6564p itself and an additional negative control, as well as an unrelated promoter of Rv0757p, were also used to promote expression of lacZ. As shown in Figure 5C, the expression of lacZ was downregulated in the Ms6564-deleted mutant M. smegmatis strains compared with the wild-type strain under all four promoters: Ms4232p, Ms4235p, Ms6806p and Ms6564p. However, there was no significant difference in the expression of lacZ between the wild-type and mutant strains when a negative control, Rv0757p, was used as promoter.
These results strongly suggested that Ms6564 can function as a positive regulator and it affected the expression of DNA damage and repair genes in M. smegmatis.
Ms6564 negatively regulates gene mutation frequencies and rates
Ms6564 was shown to bind directly to the promoter regions of many cell cycle and DNA damage and repair genes. This finding suggests that Ms6564 regulates genes necessary for DNA repair and, therefore, could indirectly affect spontaneous mutation rates and other repair processes. The gene mutation frequencies and rates in both Ms6564 overexpression and gene deletion mutant M. smegmatis strains were compared to examine this result further. Ms6564-overexpressed M. smegmatis tained a 5.1-fold lower streptomycin-resistant gene mutation frequencies (2.5 ± 0.3 × 10−9), whereas the Ms6564-deleted strain obtained 5-fold higher mutation frequency (6.06 ± 0.3 × 10−8) compared with the wild-type strains (12.8 ± 0.4 × 10−9) (Table 1). Similar results were obtained with an assay of mutation rates using a fluctuation experiment (Table 2). The P-values of the rates were calculated to be <0.05 (Table 2), indicating that these differences were statistically significant. Interestingly, when expressing the Ms6564 gene through a pMind in ΔMs6564 strain, the recombinant strain of M. smegmatis ΔMs6564/pMindD6564 re-obtained a closer frequency (14.1 ± 0.4 × 10−9) (Table 1) or mutation rates (2.6 × 10−9) (Table 2) to the wild-type strain, with no significant difference between these changes (P > 0.05). The empty pMV261 plasmid or overexpression of an unrelated gene, Ms3452, had no significant effect on mutation frequency. Therefore, these results suggest that Ms6564 regulates genes responsible for repair of spontaneous mutations in M. smegmatis mc2155.
Table 1.
The mutation frequencies of wild-type and recombinant M. smegmatis strains
| Strain | SM mutation frequency × 10−9 |
|---|---|
| Msm/WT | 12.2 ± 0.2 |
| Msm/pMV261 | 12.8 ± 0.4 |
| Msm/pMV261-Ms6564 | 2.5 ± 0.3* |
| Msm/pMV261-Ms3452 | 9.3 ± 0.2 |
| Msm Ms6564:: hyg | 60.6 ± 0.3* |
| Ms6564 complementation | 14.1 ± 0.4 |
P-values of the results were calculated by unpaired two-tailed student’s test GraphPad Prism 5. *P-values of the results were <0.05.
Table 2.
The mutation rates of wild-type and recombinant M. smegmatis strains
| Fluctuation Expt no. | Strain | No. of cultures | Nt-valuea | Lea-Coulson m-valueb | Mutation ratec |
|---|---|---|---|---|---|
| 1 | Msm/WT | 30 | 1.2 × 109 | 2.4 | 2.0 × 10−9 |
| 2 | Msm/pMV261 | 30 | 1.0 × 109 | 2.5 | 2.5 × 10−9 |
| 3 | Msm/pMV261-Ms6564 | 30 | 1.2 × 109 | 0.8 | 0.7 × 10−9* |
| 4 | Msm/pMV261-Ms3452 | 30 | 1.0 × 109 | 2.2 | 2.2 × 10−9 |
| 5 | Msm/Ms6564:: hyg | 30 | 1.1 × 109 | 7.6 | 6.9 × 10−9* |
| 6 | Ms6564 complementation | 30 | 0.8 × 109 | 2.1 | 2.6 × 10−9 |
aFinal number of cells in the culture.
bNumber of mutations per culture.
cProbability of mutation per cell per generation.
P-values of the results were calculated by unpaired two-tailed student's test GraphPad Prism 5. *P-values of the results were <0.05.
DISCUSSION
The fast-growing M. smegmatis contains a large number of regulatory factors, and it has been widely used as a model organism to study the gene regulatory mechanism of the virulent M. tuberculosis (5). In the present study, M. smegmatis Ms6564 was confirmed as a candidate for the broad regulation of gene expression including cell cycle and DNA damage/repair genes.
Some broad regulators have been reported from M. smegmatis and other bacterial species. For example, Sharon et al. (16) characterized a KstR repressor involved in regulating a total of 159 genes and in directly controlling the expression of 83 genes in M. smegmatis and 74 genes in M. tuberculosis. The motifs within the target operator for the TetR-like transcriptional factor were demonstrated to have an internal palindromic symmetry with an extra central base pair (12,16). A 19 bp-palindromic motif for specific recognition by Ms6564 was identified in the current study using DNaseI footprinting experiment combined with EMSA assays. Similar to many other transcription activators, Ms6564 bound to inverted repeats of an operator sequence upstream of and only partially overlapping the −35 promoter consensus sequence (Figure 2). An interesting finding from the present work was the identification of the binding motif for Ms6564 within 339 promoters of M. smegmatis genes or operons. These potential target genes covered a variety of gene families including cell cycle and DNA damage/repair genes, transcriptional regulators, DNA-directed RNA polymerase subunit beta’ and many transport and metabolism genes (Supplementary Table S5). Therefore, our findings suggested that Ms6564 may function as a broad regulator of many different function genes in M. smegmatis.
The regulators of the TetR family are often repressors and are widely distributed among bacteria (11,35). These proteins control genes, whose products are involved in multidrug resistance, enzymes implicated in different catabolic pathways, biosynthesis of antibiotics, osmotic stress and pathogenicity (12). In the current study, based on qRT-PCR and β-galactosidase activity analysis, Ms6564 may function as an activator different from most typical TetR-like regulators. However, the expressions of dinB and dinP were shown to be negatively regulated by Ms6564 in contrast to other genes (Figure 5A). We further compared the location of the binding site for Ms6564 in the promoters of dinB and dinP with that of other target genes. However, no obvious difference was observed (Supplementary Figure S4), thus, the mechanism for the repression or activation of dinB and dinP remains to be characterized. Interestingly, a recent study suggested that mycobacterial DinB homologs were substantially different from their E. coli counterparts and deletion of these genes did not affect bacterial growth and survival (36).
Two different mechanisms, the RecA/LexA-dependent and RecA-independent mechanisms, have been described for DNA damage repair in the bacterial Save Our Ship response. The RecA/LexA-dependent mechanism has been reported (7), but the majority of inducible DNA repair genes in mycobacteria are RecA-independent activations (10,37). In the present study, most of these cell cycle and DNA damage/repair target genes for Ms6564 were found to be DNA damage inducible (Supplementary Figure S3). When overexpressing Ms6564 in M. smegmatis, the recombinant strain had a lower mutation rate and mutation frequency compared with the wild-type strain. By contrast, the deletion of Ms6564 led to a higher mutation rate and mutation frequency of the mutant strain. Therefore, Ms6564 may play an important role in the mutagensis of M. smegmatis mc2 155. Notably, these target genes included both RecA-independent (Ms1622, Ms1943, Ms4925, Ms5451 and Ms6806) and RecA-dependent (Ms2313) DNA damage and repair genes. In a recent study, a ClpR-like transcriptional factor was characterized to bind with the RecA-independent promoter motif, RecA-NDp, and to affect expressions of RecA-independent genes (38). Interestingly, the binding site for Ms6564 in the promoter region of the target gene was closer to start code than that for the ClpR-like regulator (Supplementary Figure S5). This suggests that Ms6564 might have a different mechanism for the modulation of genes responsible for DNA damage/repair in M. smegmatis.
In summary, a TetR-like family transcriptional factor, Ms6564, in M. smegmatis, was found to be a master regulator affecting mycobacterial gene mutation rates. About 339 promoters of M. smegmatis genes or operons were characterized as its potential targets. Notably, Ms6564 was found to be involved in regulating the expressions of 37 cell cycle and DNA damage/repair genes. Mycobacterial gene mutation rates were also confirmed to correlate significantly with the expression level of Ms6564. These findings suggested that Ms6564 may function as a global regulator and may activate DNA repair genes.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online: Supplementary Tables S1–S5, Supplementary Figures S1–S5.
FUNDING
The National Natural Science Foundation of China (30930003 and 31025002), the Fundamental Research Funds for the Central Universities (2011PY140) and Hubei Chutian Scholar Program (to Z.-G.H.). Funding for open access charge: The National Natural Science Foundation of China.
Conflict of interest statement. None declared.
Supplementary Material
REFERENCES
- 1.Cole ST, Brosch R, Parkhill J, Garnier T, Churcher C, Harris D, Gordon SV, Eiglmeier K, Gas S, Barry CE, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. doi: 10.1038/31159. [DOI] [PubMed] [Google Scholar]
- 2.Guo M, Feng H, Zhang J, Wang W, Wang Y, Li Y, Gao C, Chen H, Feng Y, He ZG. Dissecting transcription regulatory pathways through a new bacterial one-hybrid reporter system. Genome Res. 2009;19:1301–1308. doi: 10.1101/gr.086595.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.World Health Organization (WHO) Geneva: World Health Organization; 2010. Global tuberculosis control WHO report 2010; pp. 5–7. [Google Scholar]
- 4.Mizrahi V, Andersen SJ. DNA repair in Mycobacterium tuberculosis. What have we learnt from the genome sequence? Mol. Microbiol. 1998;29:1331–1339. doi: 10.1046/j.1365-2958.1998.01038.x. [DOI] [PubMed] [Google Scholar]
- 5.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 6.Eisen JA, Hanawalt PC. A phylogenomic study of DNA repair genes, proteins, and processes. Mutat. Res. 1999;435:171–213. doi: 10.1016/s0921-8777(99)00050-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Davis EO, Springer B, Gopaul KK, Papavinasasundaram KG, Sander P, Böttger EC. DNA damage induction of recA in Mycobacterium tuberculosis independently of RecA and LexA. Mol. Microbiol. 2002;46:791–800. doi: 10.1046/j.1365-2958.2002.03199.x. [DOI] [PubMed] [Google Scholar]
- 8.Petit C, Cayrol C, Lesca C, Kaiser P, Thompson C, Defais M. Characterization of dinY, a new Escherichia coli DNA repair gene whose products are damage inducible even in a lexA(Def) background. J. Bacteriol. 1993;175:642–646. doi: 10.1128/jb.175.3.642-646.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kleinsteuber S, Quiñones A. Expression of the dnaB gene of Escherichia coli is inducible by replication-blocking DNA damage in a recA-independent manner. Mol. Gen. Genet. 1995;248:695–702. doi: 10.1007/BF02191709. [DOI] [PubMed] [Google Scholar]
- 10.Rand L, Hinds J, Springer B, Sander P, Buxton RS, Davis EO. The majority of inducible DNA repair genes in Mycobacterium tuberculosis are induced independently of RecA. Mol. Microbiol. 2003;50:1031–1042. doi: 10.1046/j.1365-2958.2003.03765.x. [DOI] [PubMed] [Google Scholar]
- 11.Orth P, Schnappinger D, Hillen W, Saenger W, Hinrichs W. Structural basis of gene regulation by the tetracycline inducible Tet repressor-operator system. Nat. Struct. Biol. 2000;7:215–219. doi: 10.1038/73324. [DOI] [PubMed] [Google Scholar]
- 12.Ramos JL, Martínez-Bueno M, Molina-Henares AJ, Terán W, Watanabe K, Zhang X, Gallegos MT, Brennan R, Tobes R. The TetR family of transcriptional repressors. Microbiol. Mol. Biol. Rev. 2005;69:326–356. doi: 10.1128/MMBR.69.2.326-356.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schumacher MA, Miller MC, Grkovic S, Brown MH, Skurray RA, Brennan RG. Structural mechanisms of QacR induction and multidrug recognition. Science. 2001;294:2158–2163. doi: 10.1126/science.1066020. [DOI] [PubMed] [Google Scholar]
- 14.Baulard AR, Betts JC, Engohang-Ndong J, Quan S, McAdam RA, Brennan PJ, Locht C, Besra GS. Activation of the pro-drug ethionamide is regulated in mycobacteria. J. Biol. Chem. 2000;275:28326–28331. doi: 10.1074/jbc.M003744200. [DOI] [PubMed] [Google Scholar]
- 15.Dover LG, Corsino PE, Daniels IR, Cocklin SL, Tatituri V, Besra GS, Fütterer K. Crystal structure of the TetR/CamR family repressor Mycobacterium tuberculosis EthR implicated in ethionamide resistance. J. Mol. Biol. 2004;340:1095–1105. doi: 10.1016/j.jmb.2004.06.003. [DOI] [PubMed] [Google Scholar]
- 16.Kendall SL, Withers M, Soffair CN, Moreland NJ, Gurcha S, Sidders B, Frita R, Ten Bokum A, Besra GS, Lott JS, et al. A highly conserved transcriptional repressor controls a large regulon involved in lipid degradation in Mycobacterium smegmatis and Mycobacterium tuberculosis. Mol. Microbiol. 2007;65:684–699. doi: 10.1111/j.1365-2958.2007.05827.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kloosterman TG, van der Kooi-Pol MM, Bijlsma JJE, Kuipers OP. The novel transcriptional regulator SczA mediates protection against Zn2+ stress by activation of the Zn2+-resistance gene czcD in Streptococcus pneumoniae. Mol. Microbiol. 2007;65:1049–1063. doi: 10.1111/j.1365-2958.2007.05849.x. [DOI] [PubMed] [Google Scholar]
- 18.He ZG, Rezende LF, Willcox S, Griffith JD, Richardson CC. The carboxyl-terminal domain of bacteriophage T7 single-stranded DNA-binding protein modulates DNA binding and interaction with T7 DNA polymerase. J. Biol. Chem. 2003;278:29538–29545. doi: 10.1074/jbc.M304318200. [DOI] [PubMed] [Google Scholar]
- 19.Li Y, Zeng J, Zhang H, He ZG. The characterization of conserved binding motifs and potential target genes for M. tuberculosis MtrAB reveals a link between the two-component system and the drug resistance of M. smegmatis. BMC Microbiol. 2010;10:242. doi: 10.1186/1471-2180-10-242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Snapper SB, Melton RE, Mustafa S, Kieser T, Jacobs WR. Isolation and characterization of efficient plasmid transformation mutants of Mycobacterium smegmatis. Mol. Microbiol. 1990;4:1911–1919. doi: 10.1111/j.1365-2958.1990.tb02040.x. [DOI] [PubMed] [Google Scholar]
- 21.Rodriguez GM, Voskuil MI, Gold B, Schoolnik GK, Smith I. ideR, an essential gene in mycobacterium tuberculosis: role of IdeR in iron-dependent gene expression, iron metabolism, and oxidative stress response. Infect. Immun. 2002;70:3371–3381. doi: 10.1128/IAI.70.7.3371-3381.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Blokpoel MCJ, Murphy HN, O’Toole R, Wiles S, Runn ESC, Stewart GR, Young DB, Robertson BD. Tetracycline-inducible gene regulation in mycobacteria. Nucleic Acids Res. 2005;33:e22. doi: 10.1093/nar/gni023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang M, Gao C, Wang Y, Zhang H, He ZG. Characterization of the interaction and cross-regulation of three Mycobacterium tuberculosis RelBE modules. PLoS ONE. 2010;5:e10672. doi: 10.1371/journal.pone.0010672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Möker N, Brocker M, Schaffer S, Krämer R, Morbach S, Bott M. Deletion of the genes encoding the MtrA-MtrB two-component system of Corynebacterium glutamicum has a strong influence on cell morphology, antibiotics susceptibility and expression of genes involved in osmoprotection. Mol. Microbiol. 2004;54:420–438. doi: 10.1111/j.1365-2958.2004.04249.x. [DOI] [PubMed] [Google Scholar]
- 25.Livak KJ, Schmittgen TD. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 26.Frank SR, Schroeder M, Fernandez P, Taubert S, Amati B. Binding of c-Myc to chromatin mediates mitogen-induced acetylation of histone H4 and gene activation. Genes Dev. 2001;15:2069–2082. doi: 10.1101/gad.906601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Stover CK, de la Cruz VF, Fuerst TR, Burlein JE, Benson LA, Bennett LT, Bansal GP, Young JF, Lee MH, Hatfull GF. New use of BCG for recombinant vaccines. Nature. 1991;351:456–460. doi: 10.1038/351456a0. [DOI] [PubMed] [Google Scholar]
- 28.Miller JH. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 1972. Experiments in Molecular Genetics; pp. 352–355. [Google Scholar]
- 29.Boshoff HIM, Reed MB, Barry CE, III, Mizrahi V. DnaE2 polymerase contributes to in vivo survival and the emergence of drug resistance in Mycobacterium tuberculosis. Cell. 2003;113:183–193. doi: 10.1016/s0092-8674(03)00270-8. [DOI] [PubMed] [Google Scholar]
- 30.Rosche WA, Foster PL. Determining mutation rates in bacterial populations. Methods. 2000;20:4–17. doi: 10.1006/meth.1999.0901. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Machowski EE, Barichievy S, Springer B, Durbach SI, Mizrahi V. In vitro analysis of rates and spectra of mutations in a polymorphic region of the Rv0746 PE_PGRS gene of Mycobacterium tuberculosis. J. Bacteriol. 2007;189:2190–2195. doi: 10.1128/JB.01647-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lea DE, Coulson CA. The distribution of the numbers of mutants in bacterial populations. J. Genet. 1949;49:264–285. doi: 10.1007/BF02986080. [DOI] [PubMed] [Google Scholar]
- 33.Hall BM, Ma CX, Liang P, Singh KK. Fluctuation analysis CalculatOR: a web tool for the determination of mutation rate using Luria-Delbruck fluctuation analysis. Bioinformatics. 2009;25:1564–1565. doi: 10.1093/bioinformatics/btp253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Levy SB, McMurry LM, Barbosa TM, Burdett V, Courvalin P, Hillen W, Roberts MC, Rood JI, Taylor DE. Nomenclature for new tetracycline resistance determinants. Antimicrob. Agents Chemother. 1999;43:1523–1524. doi: 10.1128/aac.43.6.1523. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kana BD, Abrahams GL, Sung N, Warner DF, Gordhan BG, Machowski EE, Tsenova L, Sacchettini JC, Stoker NG, Kaplan G, et al. Role of the DinB homologs Rv1537 and Rv3056 in Mycobacterium tuberculosis. J. Bacteriol. 2010;192:2220–2227. doi: 10.1128/JB.01135-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Brooks PC, Dawson LF, Rand L, Davis EO. The mycobacterium-specific gene Rv2719c is DNA damage inducible independently of RecA. J. Bacteriol. 2006;188:6034–6038. doi: 10.1128/JB.00340-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang Y, Huang Y, Xue C, He Y, He ZG. A ClpR-like regulator specifically recognizes a RecA-independent promoter motif and broadly regulates expression of DNA damage inducible genes in mycobacteria. J. Biol. Chem. 2011;286:31159–31167. doi: 10.1074/jbc.M111.241802. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.