Abstract
The analysis of interaction networks is crucial for understanding molecular function and has an essential impact for genomewide studies. However, the interactomes of most species are largely incomplete and computational strategies that take into account sequence homology can help compensating for this lack of information using cross-species analysis. In this work we report the construction of a porcine interactome resource. We applied sequence homology matching and carried out bi-directional BLASTp searches for the currently available protein sequence collections of human and pig. Using this homology we were able to recover, on average, 71% of the proteins annotated for human pathways for the pig. Porcine protein-protein interactions were deduced from homologous proteins with known interactions in human. The result of this work is a resource comprising 204,699 predicted porcine interactions that can be used in genome analyses in order to enhance functional interpretation of data. The data can be visualized and downloaded from http://cpdb.molgen.mpg.de/pig.
Keywords: pig protein–protein interactions, sequence homology, bioinformatics, interaction networks, genome analysis
Introduction
The pig is among the economically most important domesticated animals. Pork is the major red meat consumed worldwide1 and moreover, the pig is an important biomedical model organism for human health, particularly for understanding complex traits such as arthritis, obesity, cardiovascular disease and infection.2 Interestingly, these human diseases may be modelled better in pig than in mouse, because the pig is physiologically more similar and possibly evolutionarily more closely related to human than rodents.3 In recent years, huge efforts have been made to explore the pig genome by the generation of clone libraries, physical and radiation hybrid mapping, transcriptome and SNP microarray studies, and QTL analyses. Additionally, the Swine Genome Sequencing Consortium (SGSC) was formed in 2003 and sequencing was started within a collaborative project. A first draft of the swine genome sequence with an overall depth of 4X coverage (build 9, Sscrofa9) was finished in early 2009 and the annotated Ensembl version of this build was made available in September 2009.4,5
Multiple studies on animal health, meat quality and animal diseases have been performed in pig based on genome-wide expression data, SNP and QTL analyses and functional studies. Molecular interactions are key drivers of biological function. While for a few organisms, such as human and yeast, huge amounts of protein-protein interaction data have been generated and made publicly accessible, such resources are not available for the pig. Such studies would benefit from information on porcine functional interactions such as protein-protein interactions and molecular pathways. It has been shown in other organisms, for example in human and yeast, that such interactions are key for understanding complex cellular processes and the respective biological functions.6,7
Approach
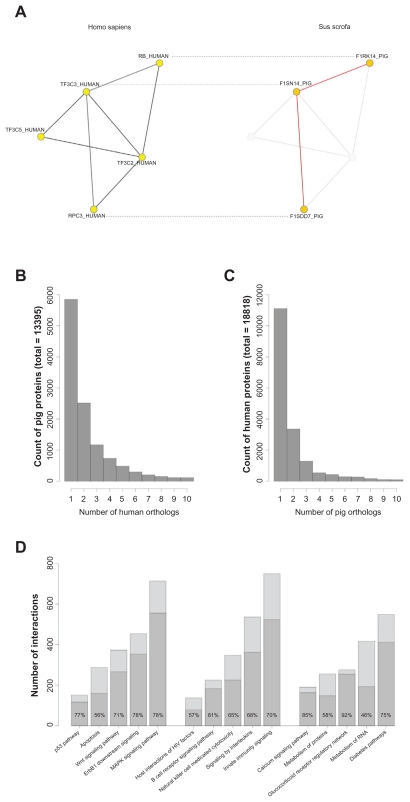
A straightforward way to extrapolate functional interactions from human to the pig is to use homology mapping of interacting proteins (Fig.1A) in order to infer porcine interactions from human interactions.8 Such homology mapping has recently been applied in order to improve genome-wide microarray probe annotations in the pig.9
Figure 1.
Homology mapping and coverage of inferred pig interactions in selected human pathways. (A) Protein-protein interaction sub-network exemplifying the concept of the pig interactome prediction. Nodes represent proteins, continuous edges represent interactions. Dashed lines connect orthologous proteins identified by sequence alignment (BLASTp). Interactions between pig proteins are inferred if a direct physical interaction is known for at least one of their human orthologs. (B) Histogram of the number of human orthologs per pig gene. The histogram is truncated on the X-axis at 10 orthologs; the maximum number is 250 (not shown). (C) Histogram of the number of pig orthologs per human gene. The histogram is truncated on the X-axis at 10 orthologs; the maximum number is 236 (not shown). (D) Coverage of predicted pig interactions in well-studied human pathways of three categories: gene regulation, immune system, and metabolism. The median percentage of orthologous pig interactions in human pathways was 71.4%.
Here, we report on the prediction of a comprehensive pig interactome using homology mapping of pig and human protein sequences. For human many different large-scale experiments, such as yeast-2-hybrid screens10 and co-immunoprecipitations,11 among others, have been performed that resulted in a large amount of protein-protein interactions. Multiple databases exist that provide free access to human molecular interactions. Recently a large number of these data resources have been agglomerated into the meta-database ConsensusPathDB,12 which currently (release 20) integrates the content of 23 human interaction resources comprising a protein-protein interaction network of 14,540 proteins and 93,292 binary interactions. Furthermore, the database contains 3,091 pre-annotated human pathways such as signaling, metabolic and gene regulatory pathways that build the basis for mapping porcine genes to known pathways.
Methods
For this study human and porcine protein sequences were downloaded from the UniProt ftp-site (ftp://ftp.uniprot.org/pub/databases/uniprot/current_release/knowledgebase/proteomes). The respective FASTAfiles contained 19,645 porcine and 72,049 human protein sequences, corresponding to 15,338 and 20,348 genes, respectively. The protein sequences were used as input data for bi-directional BLASTp (version 2.2.25) runs (pig query vs. whole-proteome human data and human query vs. whole-proteome pig data). For sequence alignment we used the BLOSUM80 substitution matrix instead of the default BLOSUM62 matrix to account for the fact that the pig genome is rather similar to the human genome. Furthermore, we used soft filtering with Smith-Waterman final alignment (the -F ‘m S’ -s T options in NCBI’s BLASTp), a parameter combination shown to result in high numbers of orthologs with low error rates.13
Post-processing of the alignment included two steps. First, we filtered matches according to the quality of the local alignments and accepted only matches with (i) BLASTp e-value <10e-10, and (ii) alignment coverage ≥90% of the input sequences, ie, the alignment between query and subject needed to cover at least 90% of their respective total lengths. Secondly, matches were kept for further analysis only if they were found in both directions of the BLASTp comparisons in order to reduce the number of false positives.
In summary, 137,904 reciprocal matches were found. On the gene level this resulted in 13,396 out of 15,338 (87.3%) different pig genes that had a human ortholog and in 18,819 out of 20,348 (92.5%) human genes that had a pig ortholog. In both cases, a majority of the genes had one or two orthologs (pig: 62.4%, human: 76.9%), and a minor proportion (pig: 14.2%, human: 9.7%) had three or four orthologs. The remaining genes (pig: 23.3%, human: 13.4%) had five or more orthologs (Fig. 1B and C).
Pathway and Interaction Mapping
By using homology mapping we were able to explore the coverage of pre-annotated human pathways with the pig orthologs and found highly conserved information. On average, human pathways contained 71% proteins with pig orthologs and, thus, these pathways build comprehensive functional information for pig studies. For example, the protein content of key biological processes including gene regulation, immune system and metabolism was conserved at rates between 46%–92% (Fig. 1D).
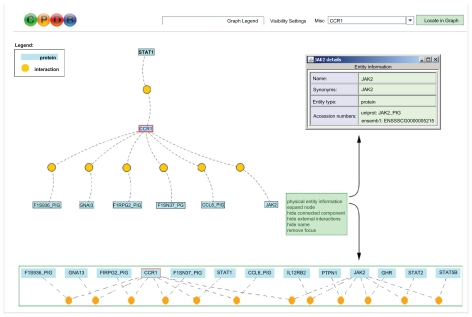
From the initial human interaction network of the ConsensusPathDB we retrieved a pig interactome consisting of 9,534 proteins and 204,699 pairwise porcine interactions. Detailed information on the interactions is given in Supplementary File 1. For each inferred binary porcine interaction it provides the orthologous human interaction. If several pig proteins were assigned to a human protein, then the corresponding interaction was listed multiple times (many-to-many relationship). Furthermore, we listed supporting publications (Pubmed identifiers) for the respective interaction, the source databases for the human interaction as well as the E-values and bit scores of the alignments of the human and porcine proteins sequences. This inferred interactome can be used for retrieving functional information from genome-wide assays, for example interactions of particular proteins found differentially expressed in microarray studies. As an example, we visualized the interaction neighbourhood of the gene CCR1 with the web interface provided at http://cpdb.molgen.mpg.de/pig (Fig. 2).
Figure 2.
Protein–protein interaction network visualization through the CPDB web server. Interaction neighbourhood of the pig gene CCR1 as retrieved and visualized from the web server. Nodes correspond to interactions and rectangles to proteins. Entity information such as gene symbols, Ensembl or UniProt identifiers can be retrieved by clicking on the respective protein node. Furthermore, the network can be expanded by clicking on individual proteins and adding some or all of its interactions; the resulting expanded network with additional JAK2 interactions is shown at the bottom of the figure.
Moreover, the E-values (or bit scores), the information on the supporting publications for the interaction and the number of different annotating databases can be used as a quality control factor in order to extract subsets of high-confidence interactions from the resource.
The top twenty pig proteins with the greatest number of predicted interactions are shown in Table 1. Among these we observed highly conserved proteins that are expressed in almost every eukaryotic cell such as proteins from the 14-3-3 family, a family consisting of seven isoforms in mammals. These proteins exert regulatory functions in key signaling pathways such as cell cycle, apoptosis, cell growth and differentiation.14
Table 1.
Top twenty pig genes sorted according to their number of interaction partners.
| Number | Porcine protein (UniProt identifier) | Human ortholog (UniProt identifier) | Number of interaction partners |
|---|---|---|---|
| 1 | F1SDS8_PIG | E7ERE7_HUMAN | 2719 |
| 2 | MYC_PIG | MYC_HUMAN | 1474 |
| 3 | F2Z558_PIG | 1433Z_HUMAN | 1141 |
| 4 | SMAD4_PIG | SMAD4_HUMAN | 1067 |
| 5 | F1SRA1_PIG | GRAP2_HUMAN | 1012 |
| 6 | F1S9F0_PIG | 1433B_HUMAN | 951 |
| 7 | TRAF6_PIG | TRAF6_HUMAN | 932 |
| 8 | F1RYA0_PIG | MK13_HUMAN | 815 |
| 9 | F1RFW6_PIG | E2F4_HUMAN | 785 |
| 10 | P53_PIG | P53_HUMAN | 744 |
| 11 | F2Z5G5_PIG | ACTZ_HUMAN | 723 |
| 12 | F2Z4Z7_PIG | ACL6B_HUMAN | 711 |
| 13 | JUN_PIG | JUN_HUMAN | 684 |
| 14 | F1SSQ9_PIG | ERBB4_HUMAN | 659 |
| 15 | F1SF01_PIG | IKKE_HUMAN | 653 |
| 16 | Q4PJJ8_PIG | F5H0K0_HUMAN | 625 |
| 17 | CDC42_PIG | CDC42_HUMAN | 599 |
| 18 | F1S1 N5_PIG | 1A02_HUMAN | 584 |
| 19 | F1S1 N1_PIG | 1A68_HUMAN | 567 |
| 20 | F1RTI0_PIG | 1A23_HUMAN | 563 |
Validation of the Inferred Pig Interactome by Whole-Genome Expression Data
A direct validation of the predicted interactions is not possible as protein–protein interaction measurements are currently rare for pig. Thus, to validate the interaction set we applied an indirect approach based on previous studies showing that interacting proteins tend to be correlated in their gene expression.15,16
In short, we obtained public whole-genome pig gene expression data from a recent study that measured expression intensities across 74 samples using Affymetrix Porcine Genome Arrays.17 Based on these data we assessed the average Pearson correlation coefficient (PCC) between the expression profiles of those pig genes whose protein products were predicted to interact. The average PCC was calculated after removing the top 5% of the pig proteins with the highest number of interaction partners (this corresponded to removing proteins with >200 interactions), since interactions of such ‘hub’ proteins are less likely to be specific.
The resulting average PCC of 0.055 was compared to a background model resulting from 1000 randomizations of the predicted interaction network through random link rewiring.18 The mean and standard deviation of the average PCC in the background model were mean = 0.024 and sd = 5.4e-5, accordingly, yielding a Z-score of 574 for the average PCC of the inferred pig interactome. This rather high Z-score confirms the consistency and relevancy of the computed porcine interactions.
Web Server and Data Access
The predicted porcine interactome can be queried and interaction modules can be visualized via the ConsensusPathDB web server (http://cpdb.molgen.mpg.de/pig). The web interface allows the search for specific pig proteins, either given as gene symbols, UniProt, Entrez or Ensembl identifiers, and the retrieval of all inferred porcine interactions of that protein. Interactions can be selected and visualized, and networks can be expanded by adding interaction partners of single proteins (Fig. 2). Furthermore, the web server can be used to search for the shortest path between two given proteins or to obtain additional information about interactions and proteins.
The complete set of interactions is given in Supplementary File 1. In addition, the set of human biological pathways with the respective pig orthologs is given in Supplementary File 2, and results of the correlation analysis regarding gene expression of interacting proteins are given in Supplementary File 3. Finally, original output files of pig vs. human and human vs. pig BLASTp runs are given in Supplementary Files 4 and 5.
Summary
Based on a comprehensive integrated human interaction network, we have compiled a predicted porcine interactome resource using homology mapping of human and pig protein sequences. The interactions can be accessed and visualized freely through a web server. The resource can be used for retrieving functional information to pig experimental studies and to conduct network-based inference.
Supplementary Data
Supplementary file 1
http://cpdb.molgen.mpg.de/download/pig_interactome_inferred.txt.gz (4.7 MB).
This table contains all inferred porcine interactions together with their human orthologs. The column description is given below:
Source_databases: databases containing the interaction
Interaction_publications: Pubmed-IDs of publications including evidence for the interaction
Nr_publications: Number of publications including evidence for the interaction
Participant_1_HSA: UniProt-ID of the first human interacting protein
Participant_2_HSA: UniProt-ID of the second human interacting protein
Participant_1_SSC: UniProt-ID of the first pig interacting protein
Participant_2_SSC: UniProt-ID of the second pig interacting protein
Evalue_1: mean E-value of bi-directional BLASTp between Participant_1_HSA and Participant_1_SSC
Evalue_2: mean E-value of bi-directional BLASTp between Participant_2_HSA and Participant_2_SSC
Bitscore_1: mean bit score of bi-directional BLASTp between Participant_1_HSA and Participant_1_SSC
Bitscore_2: mean bit score of bi-directional BLASTp between Participant_2_HSA and Participant_2_SSC.
Supplementary file 2
http://cpdb.molgen.mpg.de/download/pathways_pig_orthology.txt.gz (1.1 MB).
This table contains all inferred porcine pathways together with their human orthologs. The column description is given below:
Pathway: title of the human pathway
Source: database containing the pathway
Components (human): UniProt-IDs of the human proteins participating in the pathway
Components (pig): UniProt-IDs of the pig proteins participating in the pathway. If multiple pig orthologs were found for one human protein, these are put in parentheses. ‘---’ indicates that no pig ortholog was found for the respective human protein.
Supplementary file 3
http://cpdb.molgen.mpg.de/download/expression_correlation.txt.gz (0.1 MB).
This file contains gene expression based correlation analysis results for pig proteins that are part of the predicted interactome. The column description is given below:
Participant_1: UniProt-ID of the first pig interacting protein
Participant_2: UniProt-ID of the second pig interacting protein
Pearson_correlation_coefficient: Pearson correlation coefficient of the according two genes, whose expression was measured across 74 microarray samples.
Supplementary file 4
http://cpdb.molgen.mpg.de/download/blastpResult_pigQuery_humanTarget.txt.gz (31 MB).
This file contains the original BLASTp output of the sequence alignment between pig (query) and human (subject) proteins. The following command line options were used: -M BLOSUM80 -F “m S” -s T -m 8 -e 0.001.
Supplementary file 5
http://cpdb.molgen.mpg.de/download/blastpResult_humanQuery_pigTarget.txt.gz (58 MB).
This file contains the original BLASTp output of the sequence alignment between human (query) and pig (subject) proteins. The following command line options were used: -M BLOSUM80 -F “m S” -s T -m 8 -e 0.001.
Acknowledgements
The work was funded by the German Science Foundation (grants HE 4607/2-1 and 4607/3-1) and the BMBF Fugato-Plus program (grant REPORI 0315129D).
Footnotes
Disclosures
Author(s) have provided signed confirmations to the publisher of their compliance with all applicable legal and ethical obligations in respect to declaration of conflicts of interest, funding, authorship and contributorship, and compliance with ethical requirements in respect to treatment of human and animal test subjects. If this article contains identifiable human subject(s) author(s) were required to supply signed patient consent prior to publication. Author(s) have confirmed that the published article is unique and not under consideration nor published by any other publication and that they have consent to reproduce any copyrighted material. The peer reviewers declared no conflicts of interest.
References
- 1.Rothschild MF, Ruvinsky A. The Genetics of the Pig. CABI Press; Oxon: 1998. [Google Scholar]
- 2.Lunney JK. Advances in Swine Biomedical Model Genomics. Int J Biol Sci. 2007;3(3):179–84. doi: 10.7150/ijbs.3.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jorgensen FG, Hobolth A, Hornshoj H, Bendixen C, Fredholm M, Schierup MH. Comparative analysis of protein coding sequences from human, mouse and the domesticated pig. BMC Biol. 2005;3:2. doi: 10.1186/1741-7007-3-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Archibald AL, Bolund L, Churcher C, et al. Pig genome sequence—analysis and publication strategy. BMC Genomics. 2010;11:438. doi: 10.1186/1471-2164-11-438. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fan B, Gorbach DM, Rothschild MF. The pig genome project has plenty to squeal about. Cytogenet Genome Res. 2011;134(1):9–18. doi: 10.1159/000324043. [DOI] [PubMed] [Google Scholar]
- 6.Han JD. Understanding biological functions through molecular networks. Cell Res. 2008;18(2):224–37. doi: 10.1038/cr.2008.16. [DOI] [PubMed] [Google Scholar]
- 7.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18(4):644–52. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sharan R, Suthram S, Kelley RM, et al. Conserved patterns of protein interaction in multiple species. Proc Natl Acad Sci U S A. 2005;102(6):1974–9. doi: 10.1073/pnas.0409522102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tsai S, Cassady JP, Freking BA, Nonneman DJ, Rohrer GA, Piedrahita JA. Annotation of the Affymetrix porcine genome microarray. Anim Genet. 2006;37(4):423–4. doi: 10.1111/j.1365-2052.2006.01460.x. [DOI] [PubMed] [Google Scholar]
- 10.Fields S, Song O. A novel genetic system to detect protein-protein interactions. Nature. 1989;340(6230):245–6. doi: 10.1038/340245a0. [DOI] [PubMed] [Google Scholar]
- 11.Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. doi: 10.1038/nature01511. [DOI] [PubMed] [Google Scholar]
- 12.Kamburov A, Pentchev K, Galicka H, Wierling C, Lehrach H, Herwig R. ConsensusPathDB: toward a more complete picture of cell biology. Nucleic Acids Res. 2011;39:D712–7. doi: 10.1093/nar/gkq1156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moreno-Hagelsieb G, Latimer K. Choosing BLAST options for better detection of orthologs as reciprocal best hits. Bioinformatics. 2008;24(3):319–24. doi: 10.1093/bioinformatics/btm585. [DOI] [PubMed] [Google Scholar]
- 14.Mhawech P. 14-3-3 proteins—an update. Cell Res. 2005;15(4):228–36. doi: 10.1038/sj.cr.7290291. [DOI] [PubMed] [Google Scholar]
- 15.Ge H, Liu Z, Church GM, Vidal M. Correlation between transcriptome and interactome mapping data from Saccharomyces cerevisiae. Nat Genet. 2001;29(4):482–6. doi: 10.1038/ng776. [DOI] [PubMed] [Google Scholar]
- 16.Deane CM, Salwiński Ł, Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol Cell Proteomics. 2002;1(5):349–56. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 17.Ponsuksili S, Murani E, Schwerin M, Schellander K, Wimmers K. Identification of expression QTL (eQTL) of genes expressed in porcine M. longissimus dorsi and associated with meat quality traits. BMC Genomics. 2010;11:572. doi: 10.1186/1471-2164-11-572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Maslov S, Sneppen K, Zaliznyak A. Detection of topological patterns in complex networks: correlation profile of the internet. Physica A: Statistical Mechanics and its Applications. 2004;333:529–40. [Google Scholar]