Figure 1.
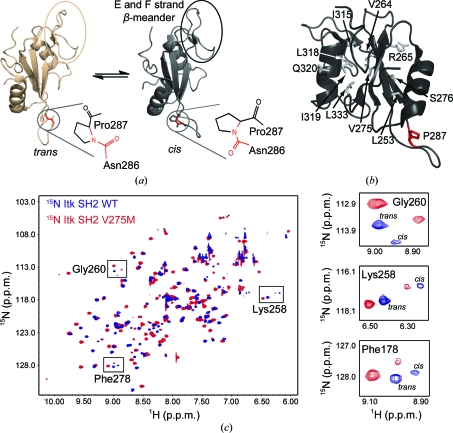
(a) Itk SH2 structures determined by NMR spectroscopy. The canonical fold of an SH2 domain consists of a central antiparallel β-sheet that is sandwiched between an N-terminal α-helix A and a C-terminal α-helix B. Pro287 (circled) interconverts between the trans and cis imide-bond conformations in solution. The trans and cis X–Pro imide bonds are shown next to each structure, with the imide bond itself indicated in red. The β-meander structure in the SH2 monomer containing the short E and F strands is circled in both structures and labeled in the cis structure. All structural figures in this report were made using PyMOL (DeLano, 2002 ▶). (b) Itk SH2 with residues targeted for mutation to methionine shown as gray sticks and labeled (Pro287 is shown in red). (c) 1H–15N HSQC spectrum of wild-type Itk SH2 (blue) overlaid with the corresponding spectrum of the Itk SH2 V275M mutant (red). Right, expanded HSQC spectra for three specific SH2 resonances, Gly260, Lys258 and Phe278, showing the cis and trans peaks for wild-type and mutant protein. The cis:trans ratio is unchanged by the mutation of Val275 to methionine.