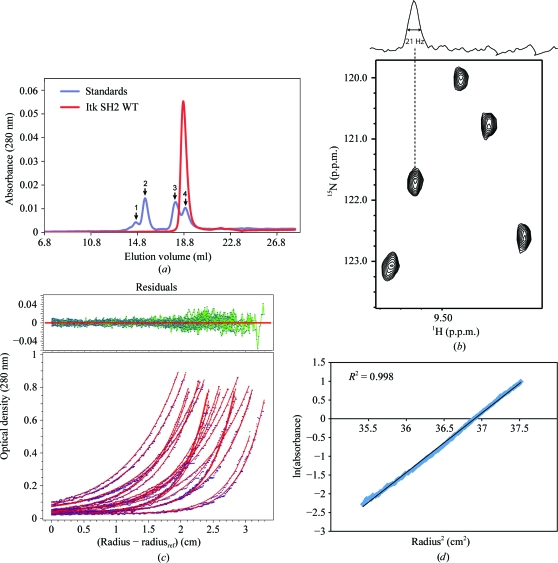
Figure 3.
(a) Size-exclusion chromatography of Itk SH2. The elution profile of Itk SH2 (red trace) is compared with the gel-filtration protein standards (blue trace). The standard peaks (1–4, indicated by arrows) correspond to albumin (65 kDa), ovalbumin (45 kDa), chymotrypsinogen A (25 kDa) and ribonuclease A (13.7 kDa) respectively. (b) NMR line-width analysis of Itk SH2. A section of the 1H–15N HSQC spectrum of Itk SH2 is shown with the one-dimensional 1H slice above. (c, d) Sedimentation-equilibrium analysis of Itk SH2. The data (three scans at 1 h interval each) were obtained at rotor speeds of 39 600, 46 400 and 56 900 rev min−1 at protein concentrations of 16, 27 and 38 µM. The solid lines (red) in (c) represent the global fit of the data to a single-component (monomer) model. Fit residuals are shown at the top. (d) Plot of the natural logarithm of the absorbance versus radius2. Only a single data set (38 µM sample spun at 46 400 rev min−1) is shown for clarity. The calculated molecular mass of Itk SH2 is 12.69 kDa, which is consistent with that of a monomeric species.