Abstract
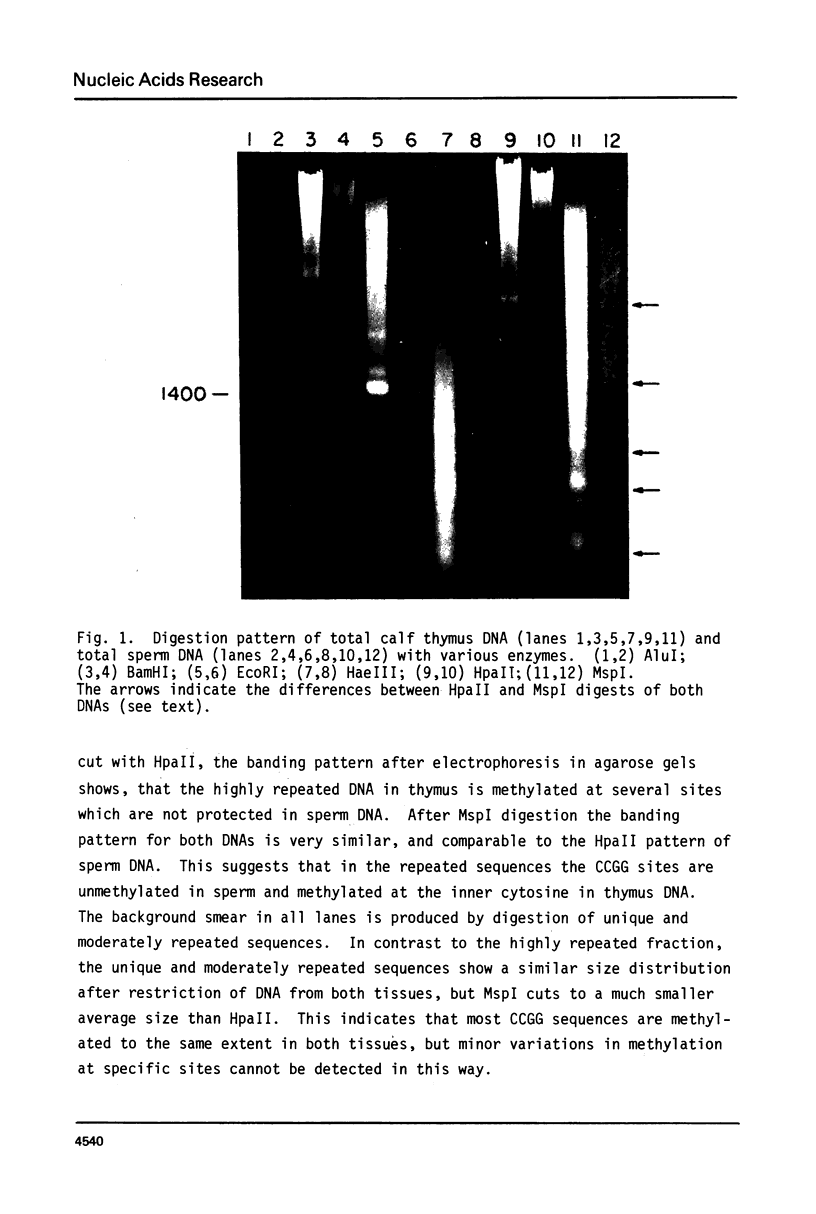
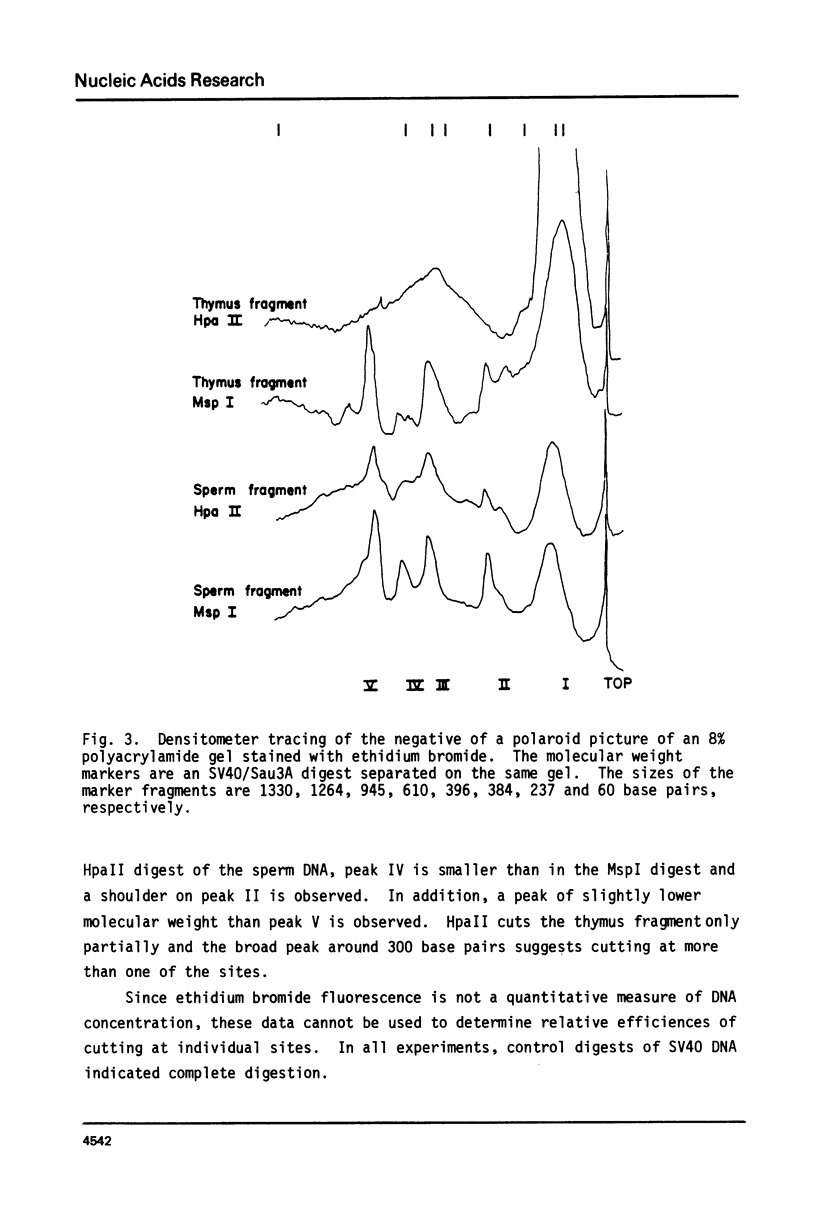
Genomic DNA of calf thymus contains 1.5 times as much 5-methylcytosine as similar sperm DNA, but the major EcoRI repeat fragment from satellite I of thymus contains ten times as much 5-methylcytosine as the corresponding fragment from sperm DNA. Restriction enzyme analyses of the total DNA and the satellite I fragment show that three HpaII sites in the fragment are completely unmethylated in sperm but fully methylated in thymus DNA. Under-methylation of many sites in the satellite DNAs can probably account for the lower level of methylation of sperm DNA rather than hemimethylation as previously suggested. These results are also discussed in relation to maintenance and de novo (initiation-type) methylases.
Full text
PDF







Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- BORENFREUND E., FITT E., BENDICH A. Isolation and properties of deoxyribonucleic acid from mammalian sperm. Nature. 1961 Sep 30;191:1375–1377. doi: 10.1038/1911375a0. [DOI] [PubMed] [Google Scholar]
- Bird A. P., Taggart M. H. Variable patterns of total DNA and rDNA methylation in animals. Nucleic Acids Res. 1980 Apr 11;8(7):1485–1497. doi: 10.1093/nar/8.7.1485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DOSKOCIL J., SORM F. Distribution of 5-methylcytosine in pyrimidine sequences of deoxyribonucleic acids. Biochim Biophys Acta. 1962 Jun 11;55:953–959. doi: 10.1016/0006-3002(62)90909-5. [DOI] [PubMed] [Google Scholar]
- Gautier F., Mayer H., Goebel W. Cloning of calf thymus satellite I DNA in Escherichia coli. Mol Gen Genet. 1976 Nov 24;149(1):23–31. doi: 10.1007/BF00275957. [DOI] [PubMed] [Google Scholar]
- Kaput J., Sneider T. W. Methylation of somatic vs germ cell DNAs analyzed by restriction endonuclease digestions. Nucleic Acids Res. 1979 Dec 20;7(8):2303–2322. doi: 10.1093/nar/7.8.2303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopecka H., Macaya G., Cortadas J., Thiéry J. P., Bernardi G. Restriction enzyme analysis of satellite DNA components from the bovine genome. Eur J Biochem. 1978 Mar;84(1):189–195. doi: 10.1111/j.1432-1033.1978.tb12156.x. [DOI] [PubMed] [Google Scholar]
- Macaya G., Cortadas J., Bernardi G. An analysis of the bovine genome by density-gradient centrifugation. Preparation of the dG+dC-rich DNA components. Eur J Biochem. 1978 Mar;84(1):179–188. doi: 10.1111/j.1432-1033.1978.tb12155.x. [DOI] [PubMed] [Google Scholar]
- Mandel J. L., Chambon P. DNA methylation: organ specific variations in the methylation pattern within and around ovalbumin and other chicken genes. Nucleic Acids Res. 1979 Dec 20;7(8):2081–2103. doi: 10.1093/nar/7.8.2081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mann M. B., Smith H. O. Specificity of Hpa II and Hae III DNA methylases. Nucleic Acids Res. 1977 Dec;4(12):4211–4221. doi: 10.1093/nar/4.12.4211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGhee J. D., Ginder G. D. Specific DNA methylation sites in the vicinity of the chicken beta-globin genes. Nature. 1979 Aug 2;280(5721):419–420. doi: 10.1038/280419a0. [DOI] [PubMed] [Google Scholar]
- Philippsen P., Streeck R. E., Zachau H. G. Investigation of the repetitive sequences in calf DNA by cleavage with restriction nucleases. Eur J Biochem. 1975 Sep 1;57(1):55–68. doi: 10.1111/j.1432-1033.1975.tb02276.x. [DOI] [PubMed] [Google Scholar]
- Pollock J. M., Jr, Swihart M., Taylor J. H. Methylation of DNA in early development: 5-methyl cytosine content of DNA in sea urchin sperm and embryos. Nucleic Acids Res. 1978 Dec;5(12):4855–4861. doi: 10.1093/nar/5.12.4855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Razin A., Riggs A. D. DNA methylation and gene function. Science. 1980 Nov 7;210(4470):604–610. doi: 10.1126/science.6254144. [DOI] [PubMed] [Google Scholar]
- Roizes G. A possible structure for calf satellite DNA I. Nucleic Acids Res. 1976 Oct;3(10):2677–2696. doi: 10.1093/nar/3.10.2677. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sneider T. W. The 5'-cytosine in CCGG1 is methylated in two eukaryotic DNAs and Msp I is sensitive to methylation at this site. Nucleic Acids Res. 1980 Sep 11;8(17):3829–3840. doi: 10.1093/nar/8.17.3829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutter D., Doerfler W. Methylation of integrated adenovirus type 12 DNA sequences in transformed cells is inversely correlated with viral gene expression. Proc Natl Acad Sci U S A. 1980 Jan;77(1):253–256. doi: 10.1073/pnas.77.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tabak H. F., Flavell R. A. A method for the recovery of DNA from agarose gels. Nucleic Acids Res. 1978 Jul;5(7):2321–2332. doi: 10.1093/nar/5.7.2321. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanyushin B. F., Tkacheva S. G., Belozersky A. N. Rare bases in animal DNA. Nature. 1970 Mar 7;225(5236):948–949. doi: 10.1038/225948a0. [DOI] [PubMed] [Google Scholar]
- Waalwijk C., Flavell R. A. DNA methylation at a CCGG sequence in the large intron of the rabbit beta-globin gene: tissue-specific variations. Nucleic Acids Res. 1978 Dec;5(12):4631–4634. doi: 10.1093/nar/5.12.4631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wigler M., Levy D., Perucho M. The somatic replication of DNA methylation. Cell. 1981 Apr;24(1):33–40. doi: 10.1016/0092-8674(81)90498-0. [DOI] [PubMed] [Google Scholar]
- van der Ploeg L. H., Flavell R. A. DNA methylation in the human gamma delta beta-globin locus in erythroid and nonerythroid tissues. Cell. 1980 Apr;19(4):947–958. doi: 10.1016/0092-8674(80)90086-0. [DOI] [PubMed] [Google Scholar]




