Abstract
For vectorborne infections, host selection by bloodfeeding arthropods dictates the interaction between host and pathogen. Because Culex mosquitoes that transmit West Nile virus (WNV) feed both on mammalian and avian hosts with varying competence, understanding the bloodfeeding patterns of these mosquitoes is important for understanding the transmission dynamics of WNV. Herein, we describe a new microsphere-based assay using Luminex xMAP® technology to rapidly identify 15 common hosts of Culex mosquitoes at our California study sites. The assay was verified with over 100 known vertebrate species samples and was used in conjunction with DNA sequencing to identify over 125 avian and mammalian host species from unknown Culex bloodmeals, more quickly and with less expense than sequencing alone. In addition, with multiplexed labeled probes, this microsphere array identified mixed bloodmeals that were difficult to discern with traditional sequencing. The microsphere set was easily expanded or reduced according to host range in a specific area, and this assay has made it possible to rapidly screen thousands of Culex spp. bloodmeals to extend our understanding of WNV transmission patterns.
Keywords: Culex, mosquito, bloodmeal, host identification, Luminex
Introduction
Host selection by bloodfeeding arthropods dictates the host-pathogen interface of arthropod-borne pathogens. This selection, along with the ability of selected hosts to produce sufficient levels of infectious pathogens, drives the efficiency of transmission and ultimately the frequency of vertebrate host infection. Understanding bloodfeeding patterns is therefore vital in understanding the transmission of zoonoses that involve multiple vertebrate hosts.
West Nile virus (WNV) is one such zoonosis, where Culex mosquito vectors feed on literally hundreds of avian as well as mammalian species (Komar 2003), including disease-susceptible horses and humans. Vertebrate species vary markedly in their host competence for WNV, as measured by their ability to become infected with WNV and achieve viral titers sufficient to infect mosquito vectors, with some vertebrate hosts producing undetectable to low viral titers and others generating serum viremias exceeding 109 infectious particles per mL. Furthermore, frequent bloodfeeding by infected vectors on non-competent or immune hosts can dampen transmission, while conversely feeding on highly competent hosts can increase the number of secondary vector infections (Keesing et al. 2006). If the preferred bloodmeal hosts are competent, they may serve as super-spreaders contributing to a high number of WNV infections despite low host abundance (Kilpatrick et al. 2006). Based on the transmission ramifications of this crucial component of arthropod-borne pathogen transmission cycles, understanding the patterns of host selection by Culex mosquitoes is vital to understanding the transmission dynamics of WNV.
The importance of delineating host selection patterns for arbovirus vectors was recognized over 60 years ago. Culex bloodmeals were first identified using serological methods (Reeves & Hammon 1944), providing a broad understanding of host selection. These methods, however, were limited in that they generally could not distinguish hosts to species level (Tempelis 1975; Washino & Tempelis 1983). Recently, several molecular techniques have been developed to genetically identify host species. These techniques include restriction fragment length polymorphism (RFLP) (Ngo & Kramer 2003; Oshaghi et al. 2006), heteroduplex analysis (Apperson et al. 2002; Lee et al. 2002; Savage et al. 2007), reverse line blot hybridization (Humair et al. 2007; Pichon et al. 2003), real-time PCR (van den Hurk et al. 2007), and most predominantly, DNA sequencing (Kent et al. 2009; Molaei et al. 2006; Molaei et al. 2010; Montgomery et al. 2011). As reviewed by Kent (2009), each of these techniques has benefits and drawbacks: RFLP assays can distinguish minor sequence differences but require a previously established profile library, heteroduplex tests can analyze many samples concurrently but may be difficult to interpret, reverse line blot hybridization is inexpensive and can screen concurrently for several hosts but throughput is limited, real-time PCR assays are high-throughput and highly specific but limited by the small number of available fluorophore labels and difficulties with multiplexing the reaction, and DNA sequencing is robust but can be time consuming and expensive as well as problematic for interpreting data from mosquitoes that imbibe multiple bloodmeals during a gonotrophic cycle.
To address some of the drawbacks of these identification methods, we describe herein a molecular assay that utilizes a microsphere array employing uniquely labeled microspheres covalently bound to oligonucleotide capture probes (Luminex xMAP®, Luminex Corporation, Austin, TX) (Dunbar 2006). These capture probes are host species-specific and hybridize to biotinylated PCR products. Using a process similar to flow cytometry, the analyzer (Luminex® 200™, Luminex Corporation, Austin, TX) interrogates each microsphere individually with two lasers. One laser identifies the microsphere, while the other quantifies the fluorescence associated with the hybridized PCR products. Samples can be identified and quantified with this platform, and up to 100 unique capture probes can be tested in one reaction (Dunbar 2006).
The purpose of the current study was to develop a high-throughput bloodmeal identification method that is less expensive than DNA sequencing, but more robust and easier to interpret than other published methods. The mitochondrial gene cytochrome c oxidase I (COI) was targeted for the development of 15 probes specific to commonly fed-upon vertebrate species. This gene is often used in DNA barcoding, a method using short genetic markers for species determination (Hebert et al. 2003; Ratnasingham & Hebert 2007), and has recently been used in mosquito bloodmeal identification (Kent et al. 2009; Montgomery et al. 2011). This novel microsphere array assay, in conjunction with DNA sequencing, could be a useful tool for rapidly identifying large numbers of Culex bloodmeals.
Materials and Methods
Known Vertebrate Samples
Blood samples from over 120 avian species, including all species for which probes were designed, were previously collected throughout California during studies of WNV seroprevalence. Blood pellets retained and stored at −80°C were used as a source of DNA for development and evaluation of the current assay. In addition, DNA from at least 5 individuals from mammalian species for which probes were designed, Bos taurus (cow), Equus caballus (horse), Capra hircus (goat), and Canis familiaris (dog), was provided by the UC Davis Veterinary Genetics Laboratory. Human DNA was isolated from field collected Culex bloodmeals previously identified by DNA sequencing.
Unknown Culex Bloodmeal Samples
Blood-engorged Culex mosquitoes were either aspirated from walk-in red boxes (Meyer 1987) or collected in CO2 (Newhouse et al. 1966) or gravid female (Cummings 1992) traps from various study sites throughout California. These samples were stored individually at −80°C until the abdomens were removed for DNA extraction.
Duration of Viable Bloodmeal DNA
Colony-reared Culex tarsalis and Culex quinquefasciatus were allowed to engorge on blood from either a House Sparrow (Passer domesticus) or Ring-necked Dove (Streptopelia capicola) and were held at 26°C or 32°C while their bloodmeals digested and eggs developed. Three mosquitoes from each group (e.g., Cx. tarsalis fed on House Sparrow and placed at 26°C) were collected at 0, 10, 15, 20, 35, 40, 45 and 60 hrs post-feeding. The extent of bloodmeal digestion was assessed, and individual mosquitoes were scored as freshly fed (Sella’s Stage II), late fed, half gravid (Sella’s Stage III–IV), sub-gravid (Sella’s Stage V), or gravid (Sella’s Stage VI–VII) (Sella 1920; WHO 1975). Mosquitoes were stored individually at −80°C prior to DNA extraction. Protocols for feeding mosquitoes on restrained House Sparrows or Ring-necked Doves were approved by the University of California Institutional Animal Use and Care Committee.
DNA Extraction
DNA was extracted from 2–3 μl of an avian blood pellet or from the entire content of individual Culex abdomens using the DNeasy® 96 Blood & Tissue Kit (Qiagen, Valencia, CA). Culex abdomens were pressed against the side of the collection tubes to expulse the bloodmeal from the mosquito gut and exoskeleton prior to lysis. The Qiagen Animal Blood Protocol was used with modification. Briefly, 20 μl of proteinase K, 200 μl of PBS, and 200 μl of Buffer AL were added to each sample. The samples were vortexed and placed on a rocking platform at 56°C for 2–4 hours. 200 μl of 100% ethanol was added and mixed thoroughly by pipetting up and down several times before samples were transferred to DNeasy® 96 Plates. After centrifugation, samples were washed once each with Qiagen Buffer AW1 and Buffer AW2 and then eluted in 150 μl of Buffer AE.
Nested PCR
A nested PCR was developed to increase the amplification of product and to reduce the possibility of including nuclear pseudogenes in the assay (Triant & Dewoody 2007). Primers were designed in the tRNA regions flanking mitochondrial gene (COI) to amplify ~1900 bp (Figure 1). Forward and reverse primers AvTrpF1 (5′-GGCCTTCAAAGCCTTAAAYAAGAGTT-3′) and AvSerR1 (5′-RRGGWWCGAYTCCTTCCTTTCTT-3′), respectively, were designed using a multiple alignment of COI from 10 complete avian mitochondrial genomes (NC_001323, NC_009736, NC_002069, NC_008547, NC_002197, NC_010094, NC_010229, NC_009684, NC_002784, NC_007678). MaTrpF1 (5′-AGACCRAGRGCCTTCAAAGCYCT-3′) and MaSerR1 (5′-BRGGRGGTTCGATTCCTTCCTT-3′) were designed using the alignment from 6 mammalian mitochondrial genomes (NC_001807, NC_001640, NC_004028, NC_002008, NC_009126, NC_001941). A mixture of these four primers was used to amplify products from all known DNA sources as well as unknown bloodmeals. Targets were PCR-amplified in volumes of 25 μl. Each reaction contained 2.5 μl 10X buffer, 2.5mM MgCl2, 250 μM of each dATP, dTTP, dCTP and dGTP, 0.6μM of each primer, 200–400 ng DNA template, 0.5 U Amplitaq® DNA Polymerase (Applied Biosystems by Life Technologies, Carlsbad, CA), and ddH2O to total reaction volume. Cycling parameters were as follows: 94°C for 5 min; 25 cycles of 94°C for 30s, 61°C for 20s, 72°C for 2 min 30s; and 72° for 5 min.
Figure 1.
Diagram of nested PCR and bloodmeal identification assay. Primers (AvTrpF1, MaTrpF1, AvSerR1, MaSerR1) flanking the mitochondrial gene COI were used to amplify ~1900bp. This amplicon was used as the template in a PCR amplifying the 658bp ‘barcoding’ region of COI, using primers VFmix (1 VF1:1 VF1d:2 VF1i) and 5′ biotinylated VRmix (1 VR1:1 VR1d:2 VR1i). Labeled PCR products were mixed with the 15 host species-specific microsphere probes (Table 1). If probe-product hybridization was detected by the Luminex200®, a positive match was made. Unidentified PCR products were sequenced using the forward primer VF1d. Sequences were subsequently identified using the Barcode of Life Data Systems (BOLD; www.boldsystems.org).
For the second step of the nested PCR (Figure 1), previously published primers and protocols (Cooper et al. 2007; Ivanova et al. 2006) were used to amplify the 658bp barcoding region of COI, using the first PCR amplicon as template. Forward primers VF1 (5′-TTCTCAACCAACCACAAAGACATTGG-3′), VF1d (5′-TTCTCAACCAACCACAARGAYATYGG-3′) and VF1i (5′-TTCTCAACCAACCAIAAIGAIATIGG-3′) were mixed at a ratio of 1 VF1: 1 VF1d: 2VF1i. Reverse primers VR1bio (5′-bio-TAGACTTCTGGGTGGCCAAAGAATCA-3′), VR1dbio (5′-bio-TAGACTTCTGGGTGGCCRAARAAYCA-3′) and VR1ibio (5′-bio-TAGACTTCTGGGTGICCIAAIAAICA-3′) were biotinylated and mixed at the same ratio 1 VR1bio: 1 VR1dbio: 2 VR1ibio. Targets were amplified using HotstarTaq Plus® DNA polymerase (Qiagen, Valencia, CA). Each reaction contained 2.5 μl 10X buffer with MgCl2, 50 μM of each dATP, dTTP, dCTP and dGTP, 0.4 μM each primer mix, 1 μl PCR product from the previous reaction, 0.5 U polymerase, and ddH2O to total 25 μl. Cycling parameters were 95°C for 5 min; 30 cycles of 95°C for 30 s, 45°C for 15 s, 72°C for 30 s; and finally 72°C for 7 min. Amplification results were verified by electrophoresis on 1.5% (w/v) agarose gel with 1X GelStar® (Lonza, Walkersville, MD) nucleic acid stain.
Probe Design
Oligonucleotide probes were designed for 15 species (10 avian and 5 mammalian) (Table 1) known to be frequently fed upon by Culex mosquitoes at our study sites based on WNV antibody seroprevalence (Wheeler et al. 2009) and/or previous bloodmeal identification studies using serological methods. The probes were designed first by using the open source software SigOli (www.lifeintel.org)(Zahariev et al. 2009). Species folders were created for each of the 15 probe species, 15 closely related species, and 25 additional species from various avian and mammalian orders. Each folder contained COI barcoding sequences for at least 10 individuals of each species. SigOli returned 12-mers that were conserved within a species folder and had at least 2 differences from every sequence outside a given folder. These 12-mers then were placed in a multiple alignment of the inclusive consensus sequences from the 50+ probe and non-probe reference species. The final 24–26bp probes were manually designed around these 12-mers to maximize differences with non-target sequences and create probes with desired thermodynamic properties. Probe specificity also was assessed using GenBank BLAST.
Table 1.
Fifteen species-specific probes developed for microsphere-based bloodmeal identification.
| Probe | Host Target | Common Name | DNA sequence (5′→3′) |
|---|---|---|---|
| amcr33 | Corvus brachyrhynchos | American Crow | ACCCTCCTTCCTTCTCCTTCTAGC |
| cogd37 | Columbina passerina | Common Ground-Dove | TCCTTATCACCGCCGTCCTCCTTC |
| wesj38 | Aphelocoma californica | Western Scrub-Jay | TGAACTGTATATCCTCCACTTGCTGG |
| bcnh42 | Nycticorax nycticorax | Black-crowned Night-Heron | CAGCCAGGAACACTACTTGGAGACG |
| greg44 | Ardea alba | Great Egret | GTCCTGATTACCGCTGTCTTACTC |
| sneg45 | Egretta thula | Snowy Egret | GCAGGTACGGGCTGAACAGTCTAC |
| modo46 | Zenaida macroura | Mourning Dove | ACCGCCGTTCTCCTCCTTCTATCC |
| hofi47 | Carpodacus mexicanus | House Finch | AGAAGCAGGGGTTGGCACAGGATG |
| hosp51 | Passer domesticus | House Sparrow | CCTATCGCTACCAGTTCTTGCTGC |
| ybma52 | Pica nuttalli | Yellow-billed Magpie | TCCTTGCCGCTGGAATTACTATGC |
| cow53 | Bostaurus | Cow | TCATTCCTACTACTCCTCGCATCC |
| horse54 | Equus caballus | Horse | AATTGAAGCAGGTGCCGGAACAGG |
| dog55 | Canis familiaris | Dog | TGGTAGAAGCAGGTGCAGGAACGG |
| goat56 | Capra hircus | Goat | TACTGCCGTACTACTCCTCCTTTC |
| human61 | Homo sapiens | Human | GCCTCCGTAGACCTAACCATCTTC |
Probes were synthesized with a 5′ amino C6 modification (Eurofins MWG Operon, Huntsville, AL) that allowed for coupling of the oligonucleotides to carboxylated Luminex MicroPlex® Microspheres using manufacturer’s protocols (www.luminexcorp.com).
Hybridization Assay
The oligonucleotide probes were used in a direct hybridization assay (Dunbar 2006) carried out in 1.5X TMAC hybridization solution [3M tetramethyl ammonium chloride, 50mM Tris-HCl (pH 8.0), 4mM EDTA (pH 8.0), 0.1% sarkosyl]. For each sample, 4 μl of biotinylated PCR product was added to 13 μl 1X Tris/EDTA buffer (pH 8.0) and 33 μl 1.5X TMAC solution containing ~5,000 microspheres coupled to each probe. Samples were denatured at 95°C for 5 min then hybridized at 58°C for 20 min, based on the predicted probe melting temperatures.. The supernatant was removed by vacuum filtration through a pre-wet 1.2 μm filter plate (Milipore, Billerica, MA). Microspheres were resuspended in 100 μl 1X TMAC solution with 0.02% Tween-20 containing 2 μg/mL streptavidin-R-phycoerythrin (PJ31S, ProZyme, Hayward, CA). Samples were held at 58°C for 5 min then analyzed at 58°C on a Luminex 200® analyzer.
DNA Sequencing and Identification
Unknown bloodmeals identified by our microsphere assay were verified by the barcoding sequence of COI. The 658bp biotinylated PCR products were treated with ExoSAP-IT® (USB) per manufacturer’s instructions to remove unwanted dNTPs and primers, and the purified products were sent for capillary array sequencing to the College of Agricultural and Environmental Sciences Genomics Facility (CGF) at UC Davis. Primers VF1d and VR1d were used in the sequencing reactions. Sequences were identified with the Barcode of Life Datasystems (BOLD) Identification Engine (www.boldsystems.org) (Kent et al. 2009a; Ratnasingham & Hebert 2007).
Results
Nested PCR
The nested PCR amplified target DNA for 130 avian and mammalian species (Figure 2; Supp Table 1) from both known vertebrate sources and unknown Culex bloodmeals. The biotinylated second PCR product was used successfully for both the microsphere assay and for identification by DNA sequencing. The PCR was tested against 107 known avian and mammalian species, all of which were successfully amplified and identified correctly. As the primers for the first PCR were designed using only avian and mammalian sequences, reptile and amphibian samples were not extensively tested. Western fence lizard (Sceloporus occidentalis) and California slender salamander (Batrachoseps attenuates) were successfully amplified and identified by sequencing, but other taxa may have been missed. Over 85% of ~3,500 unknown bloodmeals tested to date have been identified by either the microsphere assay (~75% of those identified) or DNA sequencing. Although it is possible that some proportion of those not identified was due to a lack of sensitivity of the nested PCR, most likely errors were in upstream processes such as sampling, handling and extraction.
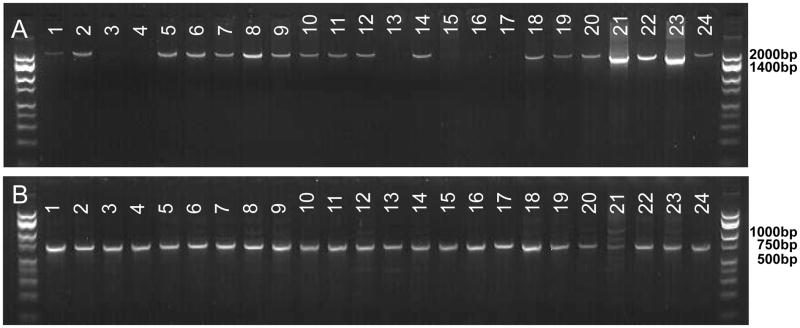
Figure 2.
(A) Product from PCR using primers flanking COI for 19 avian and 5 mammalian species. This product was used as the template for the PCR shown in B. (B) Region of COI that will be sequenced and used for microsphere assay. Lane 1: Western Scrub-Jay, 2:Yellow-billed Magpie, 3:American Crow, 4:House Sparrow, 5:Song Sparrow, 6:White-crowned Sparrow, 7:American Robin, 8:House Finch, 9:Nuttail’s Woodpecker, 10:Red-winged Blackbird, 11:Cooper’s Hawk, 12:Black-crowned Night-Heron, 13:Cattle Egret, 14:Snowy Egret, 15:California Quail, 16:Chicken, 17:Mourning Dove, 18:Common Ground-Dove, 19:Mallard, 20:Dog, 21:Cat, 22:Horse, 23:Sheep, 24:Human
Duration of Viable Bloodmeal DNA
There was no apparent difference in the rate of bloodmeal digestion between Cx. tarsalis or Cx. quinquefasciatus regardless of the avian host species. Bloodmeals were digested slightly faster at a higher temperature, with bloodfed Culex maintained at 32°C becoming gravid at least 5hr before those maintained at 26°C. Regardless of mosquito species, host species or temperature, bloodmeal DNA was successfully amplified in all freshly fed (abdomen with bright red blood), late fed (dark red blood occupying most of the abdomen) and half gravid (dark red blood occupying at least half of the abdomen) individuals. Successful amplification also occurred in all but one of 15 sub-gravid (bloodmeal digested and greatly reduced, with eggs occupying most of the abdomen) individuals, and bloodmeal DNA could even be amplified from some gravid individuals when only a trace of digested blood was still present. Although the viability of DNA from mammalian blood was not tested in this time series, several mammalian hosts were identified from half gravid and sub-gravid Culex collected in the field (data not shown). These data indicated that mosquitoes could be used in this assay up to 3 days after bloodfeeding on either avian or mammalian hosts, depending upon temperature and the retention of bloodmeal remnant in the gut.
Probe Validation
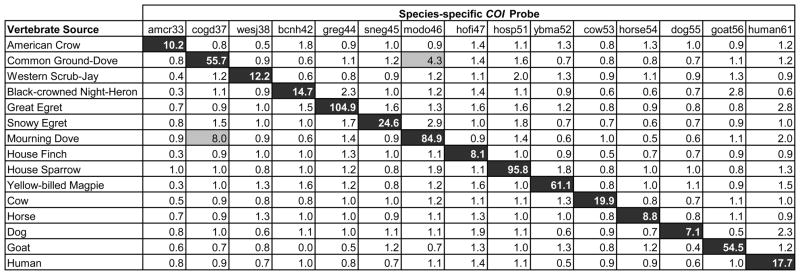
Probes were successfully developed for 15 species considered to be frequent bloodmeal hosts for Culex at our study areas (Table 1). The probes were validated against 10 – 50 individuals of each species. Probes were considered specific and were included in the assay when their ‘true-match’ median fluorescence intensity (MFI) was greater than 5 times background (established using the PCR negative control) and no non-specific hybridization resulted in an MFI >20% of the true-match. All 15 probes shown in Table 1 met these criteria (Figure 3). Some non-specific hybridization did occur among species within the same order, such as the Mourning Dove and Common Ground-Dove; however, the MFI did not cross the 20% threshold (Figure 3).
Figure 3.
Ratio of sample median fluorescence intensity (MFI) to background MFI of 15 species-specific microsphere-attached probes tested against known vertebrate DNA sources and measured by the Luminex® 200. 200ng PCR product was added to each reaction, and fluorescence was measured for 100 microspheres of each probe to determine the median value. All highlighted cells have an MFI of >5X the background MFI. Black cells represent ‘true’ matches, and gray cells show non-specific hybridization.
In addition to these 15 species, the probe set was tested against ~125 other vertebrate species. In nearly all cases, cross-hybridization was not detected. The following were exceptions. Chicken DNA yielded an approximately 50% MFI for probes amcr33 and cow53, designed for American Crow and cow, respectively. Over 50 Culex bloodmeals have produced these results, and all have been verified as chicken by DNA sequencing. The Great Egret probe (greg44) cross-reacted with a subset of Black-crowned Night-Herons with 20–30% MFI, but the Black-crowned Night-Heron probe (bcnh42) still identified these samples at over 5X the background MFI. Finally, the probe for Western Scrub-Jay (wesj38) reacted with 50% MFI to Black-headed Grosbeak (Pheucticus melanocephalus).
Because it was impossible to validate the probes against all possible vertebrate hosts, 10% of the unknown bloodmeals (~350) identified by the microsphere assay were sequenced for verification. Only three bloodmeals (<1%), one from Inca Dove (Columbina inca) and two from Black-headed Grosbeak (Pheucticus melanocephalus), were misidentified by the microsphere assay.
Mixed Bloodmeals
The microsphere assay also successfully distinguished mixed bloodmeals. When PCR products from Yellow-billed Magpie and House Sparrow were mixed after amplification, probes could detect both PCR products in ratios as low as 1:8 (Figure 4). Mixed bloodmeals also were identified from several field-caught Culex mosquitoes. These mixes were verified by looking for double peaks in the sequencing chromatograms. An example of a field-collected mixed bloodmeal is shown in Figure 5. The PCR products of this and two other mixed bloodmeals were cloned, sequenced, and the mixture of the identified hosts verified.
Figure 4.
Median Fluorescence Intensity (MFI) (with background MFI subtracted) of 100 microsphere-attached probes measured by the Luminex 200®. Known House Sparrow (HOSP) and Yellow-billed Magpie (YBMA) PCR products were mixed in the amounts given on the x-axis.
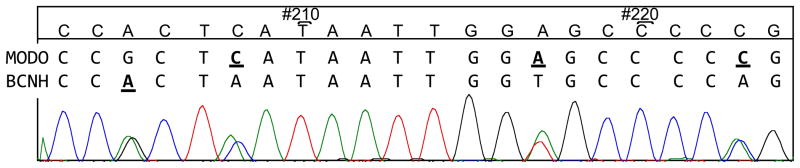
Figure 5.
Section of mixed bloodmeal sequence from a wild caught Culex mosquito. This sample was identified as blood derived from a Mourning Dove (MODO) and Black-crowned Night-Heron (BCNH) by the microsphere assay. Neither host species was identified by DNA sequencing (consensus sequence given at top) due to the roughly equal chromatogram peaks for each species. The PCR product from this sample was cloned, sequenced and verified as a mixture of MODO and BCNH.
Discussion
Molecular methods have advanced the knowledge of host selection behavior by allowing the identification of the bloodmeal host to the species level. The microsphere assay described herein adds to available methodologies by allowing for the rapid identification of the most commonly fed upon species in an area. For future work in other geographies and environments, further probes for a suite of relevant hosts could be easily added to or substituted into the current method.
This assay has major benefits in rapidly analyzing large numbers of bloodmeals. The assay is easy to interpret, because the output is numeric and no analysis of electrophoretic gels is required. It is more robust and flexible than real-time PCR with up to100 possible unique microspheres and fewer complications with multiplexing. It is less expensive and time intensive than DNA sequencing. With current prices at the UC Davis CGF, purification and sequencing of PCR products cost approximately $3.75 per sample. Using the 15-probe set described here, the cost of the microsphere assay is approximately $2.50 per sample. These prices do not include the initial PCR steps that are equivalent for both methods and, for laboratories without current access to a Luminex® system, the price does not include the initial purchase of the instrument
Further cost savings and a major benefit of the assay come from its flexibility. Because the probe mixture is made on a per plate basis, probes can be added or removed depending on host availability in a given area. For the current example, probes can be used in any combination from 1-bead set all the way up to the complete 15-bead set. In most bloodmeal identification studies, a small number of species have accounted for a large percentage of the bloodmeals. For example, in Shelby County, Tennessee, 4 species accounted for 77% of the avian feeds by the Culex pipiens complex (Savage et al. 2007), and in Weld County, Colorado, 6 host species accounted for just over 90% of the bloodmeals in July 2007 (Kent et al. 2009a). In the latter example, a 6-bead set plus 10% sequencing would cost approximately $1.22, less than 1/3 the cost of sequencing an entire plate. Because the same biotin-labeled PCR product used for the microsphere assay can be purified and sequenced, there would be no extra cost or time for additional PCR steps.
In addition to the supply cost savings, much less time is required to analyze the output data from this microsphere assay. Sequence analysis and identification can take 2–3 hours for each 96-well plate, whereas less than 15 min was required for analysis of the microsphere assay. Further benefits over sequencing include the ability to easily identify mixed bloodmeals without an additional cloning step, and the ability to quantify single or mixed bloodmeals in studies where the amount of blood taken by the vector may play an important role.
Although this assay proved cost-effective and high-throughput, there were minor drawbacks. Probe melting temperature, and thus probe length and composition, was limited in the direct hybridization assay analyzed on the Luminex 200®. The available streptavidin-RPE reporters were not thermostable at high temperatures for long time periods and high temperatures led to evaporation of the sample. Therefore, with long probes and corresponding high hybridization temperatures, background fluorescence at the end of the run was sometimes too high for analysis. The current protocol, with a hybridization temperature of 58°C and a run length of approximately 50 min, was near the limit of thermostability. Issues resulting from high hybridization temperature might be mitigated or avoided with one or more of the following adjustments: an increased volume of reporter solution could be added to extend evaporation time; on instruments with a piercing sample probe, the plate can be covered to reduce evaporation; a more thermostable reporter could be used; and/or Luminex xTAG® protocols could be used rather than the direct hybridization (xMAP®) protocols used here.
Similar to other non-sequencing methods, an inherent difficulty was that it was impossible to test our probes against all possible vertebrate host species. We evaluated our probes against all common species within our study areas, but there is always a chance for misidentification with an unexpected host. Therefore in the development stages, 10% of microsphere identifications from all study areas were sequenced to confirm that they were a true match and not a rare or unexpected host. In this manner, the current probes were tested against nearly 130 possible hosts collected in our study sites, but there may be closely related hosts in other areas that result in false positives with the current probe set.
Despite these minor drawbacks, we feel that we have provided a new and useful tool to identify blood meal hosts fed upon by Culex mosquitoes that also should be useful for other bloodfeeding arthropods. It is likely not practical to develop microsphere capture probes to identify every possible host species in a given area. However, when used in conjunction with sequencing, the assay is as robust as sequencing while being higher throughput, more efficient to interpret and less expensive than sequencing alone. Bloodmeal identification studies may still be limited by the ability to find and collect large numbers of bloodfed mosquitoes, but with using this assay, less time and money will be required to analyze those samples once they are collected.
Supplementary Material
Acknowledgments
We would like to thank Jay Well, Lisa Goldberg, Brian Carroll and Stan Langevin for their support and assistance with assay development and Niels Pedersen, UC Davis School of Veterinary Medicine, for use of laboratory space. Funding for this work was provided by grants from the Biodefense Advanced Research and Development Authority (BARDA) and the NIH Pacific Southwest Regional Center for Excellence (U54 AI065359) to AC Brault, and by grants from the California Mosquito and Vector Control Association Research Foundation and the Coachella Valley Mosquito and Vector Control Association to WK Reisen. TC Thiemann also was supported by a William Hazeltine Student Research Fellowship through the UC Davis Department of Entomology. WK Reisen acknowledges support from the Research and Policy for Infectious Disease Dynamics (RAPIDD) program of the Science & Technology Directorate, Department of Homeland Security and Fogarty International Center, National Institutes of Health.
Footnotes
Data Accessibility:
DNA sequences from vertebrate samples of known identity were deposited in GenBank (JN850661-JN850781). Supporting information for these sequences and sequence data from unknown mosquito bloodmeals was deposited in the Dryad repository: doi:10.5061/dryad.f29679q5
References
- Apperson CS, Harrison BA, Unnasch TR, et al. Host-feeding habits of Culex and other mosquitoes (Diptera: Culicidae) in the Borough of Queens in New York City, with characters and techniques for identification of Culex mosquitoes. J Med Entomol. 2002;39:777–785. doi: 10.1603/0022-2585-39.5.777. [DOI] [PubMed] [Google Scholar]
- Cooper JK, Sykes G, King S, et al. Species identification in cell culture: a two-pronged molecular approach. In Vitro Cellular & Developmental Biology-Animal. 2007;43:344–351. doi: 10.1007/s11626-007-9060-2. [DOI] [PubMed] [Google Scholar]
- Cummings RF. Design and use of a modified Reiter gravid mosquito trap for mosquito-borne encephalitis surveillance in Los Angeles County, California. Proc Mosq Vector Control Assoc Calif. 1992;60:170–176. [Google Scholar]
- Dunbar SA. Applications of Luminex xMAP technology for rapid, high-throughput multiplexed nucleic acid detection. Clin Chim Acta. 2006;363:71–82. doi: 10.1016/j.cccn.2005.06.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hebert PDN, Ratnasingham S, deWaard JR. Barcoding animal life: cytochrome c oxidase subunit 1 divergences among closely related species. P Roy Soc Lond B Bio. 2003;270:S96–S99. doi: 10.1098/rsbl.2003.0025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Humair PF, Douet V, Cadenas FM, et al. Molecular identification of bloodmeal source in Ixodes ricinus ticks using 12S rDNA as a genetic marker. J Med Entomol. 2007;44:869–880. doi: 10.1603/0022-2585(2007)44[869:miobsi]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Ivanova NV, Dewaard JR, Hebert PDN. An inexpensive, automation-friendly protocol for recovering high-quality DNA. Mol Ecology Notes. 2006;6:998–1002. [Google Scholar]
- Keesing F, Holt RD, Ostfeld RS. Effects of species diversity on disease risk. Ecology Letters. 2006;9:485–498. doi: 10.1111/j.1461-0248.2006.00885.x. [DOI] [PubMed] [Google Scholar]
- Kent RJ. Molecular methods for arthropod bloodmeal identification and applications to ecological and vector-borne disease studies. Mol Ecol Resour. 2009;9:4–18. doi: 10.1111/j.1755-0998.2008.02469.x. [DOI] [PubMed] [Google Scholar]
- Kent RJ, Juliusson L, Weissmann M, Evans S, Komar N. Seasonal blood-feeding behavior of Culex tarsalis (Diptera: Culicidae) in Weld County, Colorado, 2007. J Med Entomol. 2009;46:380–390. doi: 10.1603/033.046.0226. [DOI] [PubMed] [Google Scholar]
- Kilpatrick AM, Daszak P, Jones MJ, Marra PP, Kramer LD. Host heterogeneity dominates West Nile virus transmission. P Royal Soc B-Biol Sci. 2006;273:2327–2333. doi: 10.1098/rspb.2006.3575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komar N. West Nile virus: epidemiology and ecology in North America. Adv Virus Res. 2003;61:185–234. doi: 10.1016/s0065-3527(03)61005-5. [DOI] [PubMed] [Google Scholar]
- Lee JH, Hassan H, Hill G, et al. Identification of mosquito avian-derived blood meals by polymerase chain reaction-heteroduplex analysis. Am J Trop Med Hyg. 2002;66:599–604. doi: 10.4269/ajtmh.2002.66.599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyer RP. The “walk-in” type red box for sampling resting adult mosquitoes. Proc NJ Mosq Control Assoc. 1987;72 (1985):104. [Google Scholar]
- Molaei G, Andreadis TA, Armstrong PM, Anderson JF, Vossbrinck CR. Host feeding patterns of Culex mosquitoes and West Nile virus transmission, northeastern United States. Emerg Infect Dis. 2006;12:468–474. doi: 10.3201/eid1203.051004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molaei G, Cummings RF, Su T, et al. Vector-host interactions governing epidemiology of West Nile virus in Southern California. Am J Trop Med Hyg. 2010;83:1269–1282. doi: 10.4269/ajtmh.2010.10-0392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Montgomery MJ, Thiemann TC, Macedo P, Brown DA, Scott TW. Blood-Feeding Patterns of the Culex pipiens Complex in Sacramento and Yolo Counties, California. J Med Entomol. 2011;48:398–404. doi: 10.1603/me10067. [DOI] [PubMed] [Google Scholar]
- Newhouse VF, Chamberlain RW, Johnston JG, Jr, Sudia WD. Use of dry ice to increase mosquito catches of the CDC miniature light trap. Mosq News. 1966;26:30–35. [Google Scholar]
- Ngo KA, Kramer LD. Identification of mosquito bloodmeals using polymerase chain reaction (PCR) with order-specific primers. J Med Entomol. 2003;40:215–222. doi: 10.1603/0022-2585-40.2.215. [DOI] [PubMed] [Google Scholar]
- Oshaghi MA, Chavshin AR, Vatandoost H. Analysis of mosquito bloodmeals using RFLP markers. Exp Parasitol. 2006;114:259–264. doi: 10.1016/j.exppara.2006.04.001. [DOI] [PubMed] [Google Scholar]
- Pichon B, Egan D, Rogers M, Gray J. Detection and identification of pathogens and host DNA in unfed host-seeking Ixodes ricinus L. (Acari: Ixodidae) J Med Entomol. 2003;40:723–731. doi: 10.1603/0022-2585-40.5.723. [DOI] [PubMed] [Google Scholar]
- Ratnasingham S, Hebert PDN. BOLD: The Barcode of Life Data System ( www.barcodinglife.org) Mol Ecology Notes. 2007;7:355–364. doi: 10.1111/j.1471-8286.2007.01678.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reeves WC, Hammon WM. Feeding habits of the proven and possible mosquito vectors of western equine and St. Louis encephalitis in the Yakima Valley, Washington. Am J Trop Med. 1944;24:131–134. [Google Scholar]
- Savage HM, Aggarwal D, Apperson CS, et al. Host choice and West Nile virus infection rates in blood-fed mosquitoes, including members of the Culex pipiens complex, from Memphis and Shelby County, Tennessee, 2002–2003. Vector-Borne Zoonotic Dis. 2007;7:365–386. doi: 10.1089/vbz.2006.0602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sella M. Relazione della campagna antianofelica di Fiumicino (1919) con speciale riguardo alla biologia degli Anofeli ed agli Anofeli infetti. In: Detinova TS, editor. Ann Igiene. Suppl 85. Vol. 30. 1920. pp. 1962–1216. [Google Scholar]
- Tempelis CH. Host-feeding patterns of mosquitoes, with a review of advances in analysis of blood meals by serology. J Med Entomol. 1975;11:635–653. doi: 10.1093/jmedent/11.6.635. [DOI] [PubMed] [Google Scholar]
- Triant DA, Dewoody JA. The occurrence, detection, and avoidance of mitochondrial DNA translocations in mammalian systematics and phylogeography. Journal of Mammalogy. 2007;88:908–920. [Google Scholar]
- van den Hurk AF, Smith IL, Smith GA. Development and evaluation of real-time polymerase chain reaction assays to identify mosquito (Diptera: Culicidae) bloodmeals originating from native Australian mammals. J Med Entomol. 2007;44:85–92. doi: 10.1603/0022-2585(2007)44[85:daeorp]2.0.co;2. [DOI] [PubMed] [Google Scholar]
- Washino RK, Tempelis CH. Mosquito host-meal identification: Methodology and data analysis. Annu Rev Entomol. 1983;28:179–201. doi: 10.1146/annurev.en.28.010183.001143. [DOI] [PubMed] [Google Scholar]
- Wheeler SS, Barker CM, Fang Y, et al. Differential impact of West Nile virus on California birds. Condor. 2009;111:1–20. doi: 10.1525/cond.2009.080013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- WHO. Methods and techniques. Part II. World Health Organization; Geneva, Switzerland: 1975. Practical methods in malaria. [Google Scholar]
- Zahariev M, Dahl V, Chen W, Levesque CA. Efficient algorithms for the discovery of DNA oligonucleotide barcodes from sequence databases. Mol Ecol Resour. 2009;9:58–64. doi: 10.1111/j.1755-0998.2009.02651.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.