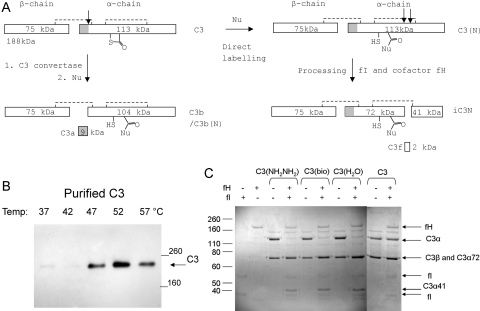
Fig. 2.
Chemical labelling of C3 thioester bond. (A) Schematic representation of C3 polypeptide structure and primary activation products. Interchain disulfide bonds are represented by dotted lines, C3a in grey and product sizes are indicated. Protease activation of C3 by C3 convertases followed by a rapid reaction of the activated thioester to surrounding nucleophilic groups to form C3b bound nucleophile C3b(N). The labelling of C3 by the direct reaction of nucleophile with the thioester without protease C3a cleavage gives C3(N). FI and cofactor fH process C3b to iC3b by protease cleavage. The C3(N) conjugate possesses a C3b like structure and can be processed in the same way (Law and Dodds, 1997). (B) Temperature dependence of biotin–PEG4–hydrazide labelling of purified C3. Purified C3 incubated with biotin–PEG4–hydrazide for 1 h at indicated temperatures. Samples analysed by 6% SDS-PAGE by Western blots of SA-HRP. (C) SDS-PAGE for comparison of C3(N) processing with fI and fH: C3(NH2NH2) C3(bio), C3(H2O) and C3. C3-conjugates were prepared by incubation with nucleophile at 52 °C for 1 h and dialysed in PBS before reaction with fI and fH for 1 h at rt. Processing of C3 detected by Coomassie staining.