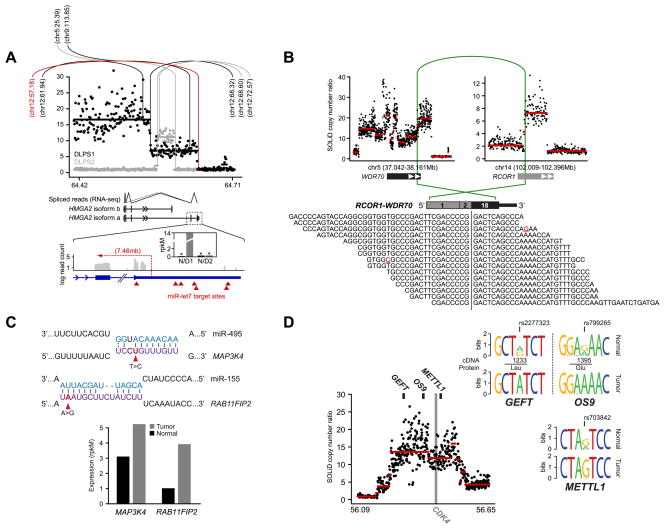
Figure 2. Diverse abnormalities manifest in RNA.
A. The segmented copy number profile, inferred from whole-genome sequencing, of the HMGA2 locus (12q14.3) is plotted for the both DLPS1 (black) and DLPS2 (gray). Rearrangements are indicated (vertical/arc lines) and the position of their second breakpoint is annotated as megabases on the recipient chromosome. In red is the structural rearrangement in DLPS1 affecting the boundary between HMGA2 exon 5 and the 3’ UTR. Splice junction reads from RNA sequencing of DLPS1 (bottom panel) confirm that the long isoform (a) is expressed. A high-resolution view of the final exons and 3’ UTR of the transcript is shown; transcriptome read coverage confirms truncation and loss of the 3’ UTR from the rearrangement and significant over-expression of HMGA2 in DLPS1 (inset; gray and black are normal and tumor samples respectively). B. Rearrangement in DLPS2 pairing a simple intragenic amplicon in RCOR1 with a complex amplicon spanning WDR70. The RCOR1 intron 2 breakpoint juxtaposed the 5’ region including exon 2 with exon 18 and the amplified 3’ end of WDR70 (green arc). In total, 15 RNA sequencing reads supported the predicted fusion junction of this out-of-frame chimera. C. Somatic mutations in the 3’ UTRs of MAP3K4 and RAB11FIP2 in DLPS2 that were detected from RNA but not exome sequencing fall in the seed regions of conserved target sites complementary to miR-495 and miR-155 respectively (purple, microRNA sequences are shown above in blue). Both genes have elevated expression in DLPS2 compared to their matched normal tissue (bottom, measured as rpkM), consonant with release from microRNA repression. D. Allele-specific expression of GEFT, OS9, and METTL1, three genes adjacent to the CDK4 oncogene on 12q13.3–14.1, was detected from heterozygous exonic variants from RNA sequencing in DLPS1 (reflected as sequence logos inferred from spanning reads) and is attributable to a single complex genomic amplification.