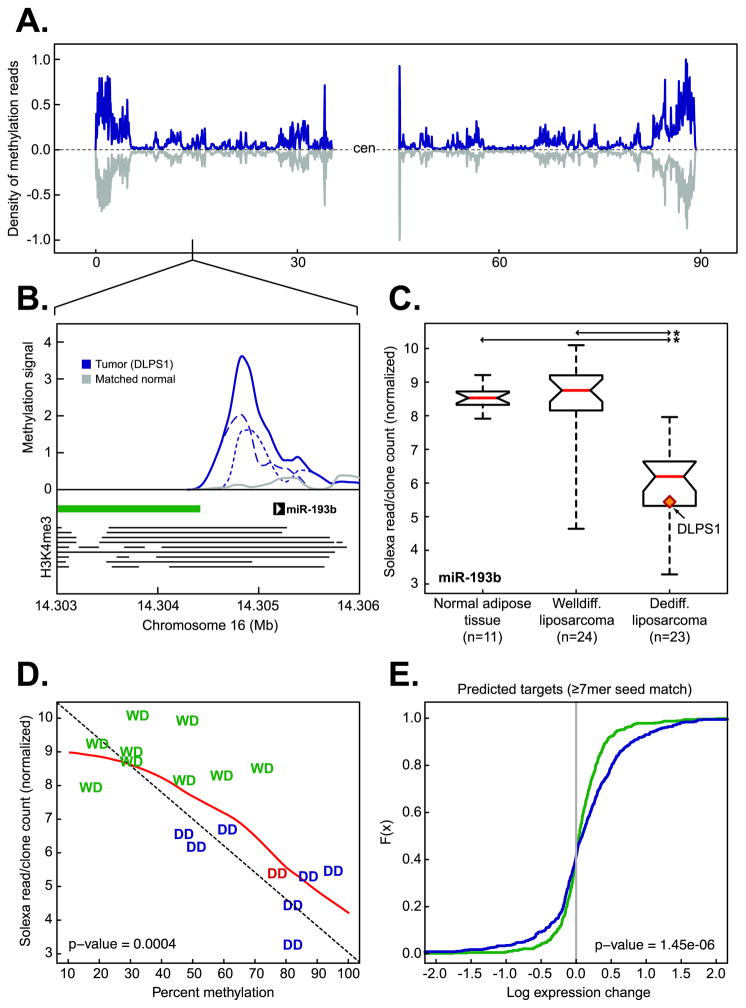
Figure 4. Epigenetic regulation of miR-193b in liposarcomagenesis.
A. The density of methylation on chromosome 16 in DLPS1 (blue) and its matched normal adipose tissue (gray). B. Methylation in the primary tumor (solid blue, positive and negative strands are dotted and dashed respectively) and matched normal adipose tissue (gray) in the region of the putative promoter [the position of enrichment of histone H3K4me3 in nine human cell types (20) is indicated by horizontal lines] of miR-193b overlapping the shore of a CpG island (green bar). C. Expression of miR-193b in normal adipose tissue samples, WLPS, and DLPS tumors determined by small RNA sequencing. The tumor in which methylation was observed (panel B) is highlighted (starred; p-value < 10−9, Student’s t-test). D. Expression of miR-193b as a function of percent methylation in the putative miR-193b promoter in a panel of both well-differentiated (WD, green) and dedifferentiated (DD, blue; DLPS1 is in red ) liposarcomas [n = 17; the diagonal is indicated by dotted line; the red line is the regression (loess)]. E. Cumulative distributions of expression changes of predicted miR-193b target genes (≥ 7 bp seed match length, n = 547 genes with expression data) between DLPS tumors and normal adipose tissues (green) or WLPS tumors and normal adipose tissues (blue) indicate that a greater number of predicted targets had increased expression in DLPS tumors with methylated miR-193b than expected by chance (p-value as indicated, Kolmogorov-Smirnov).