Abstract
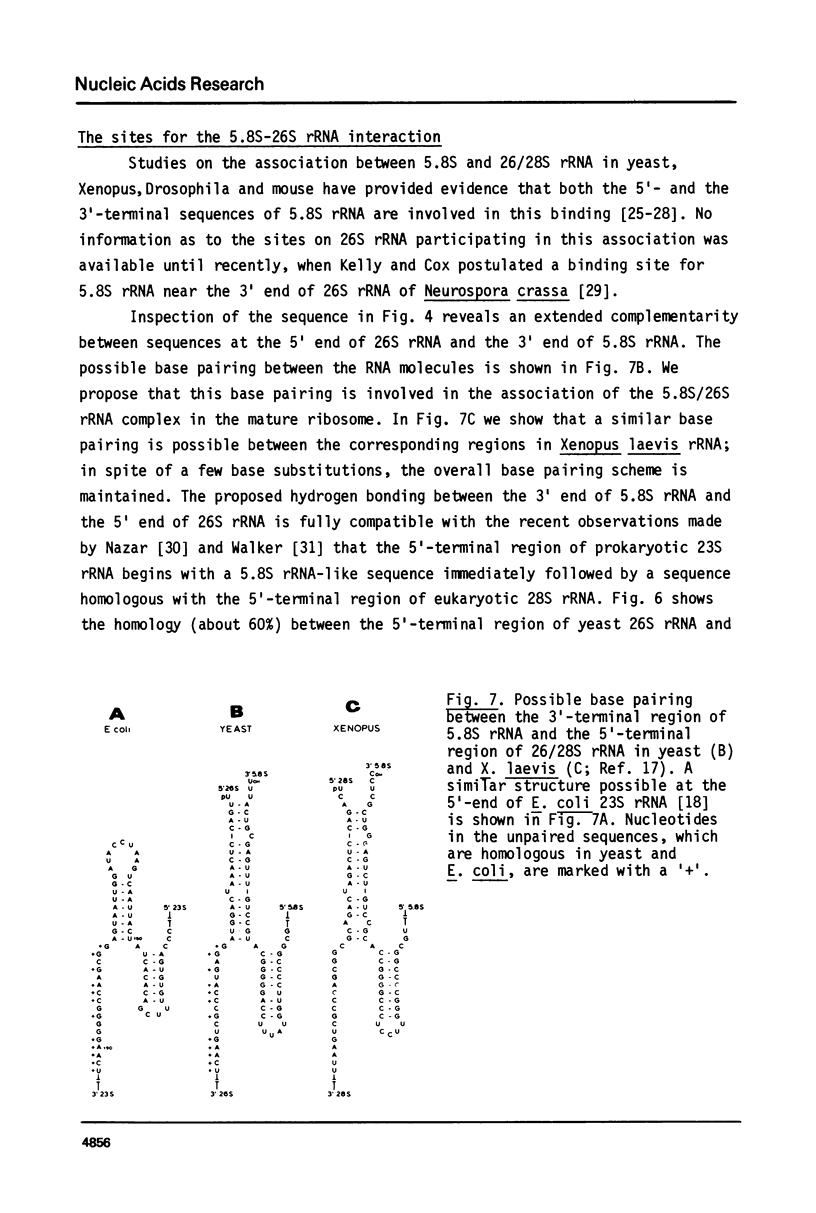
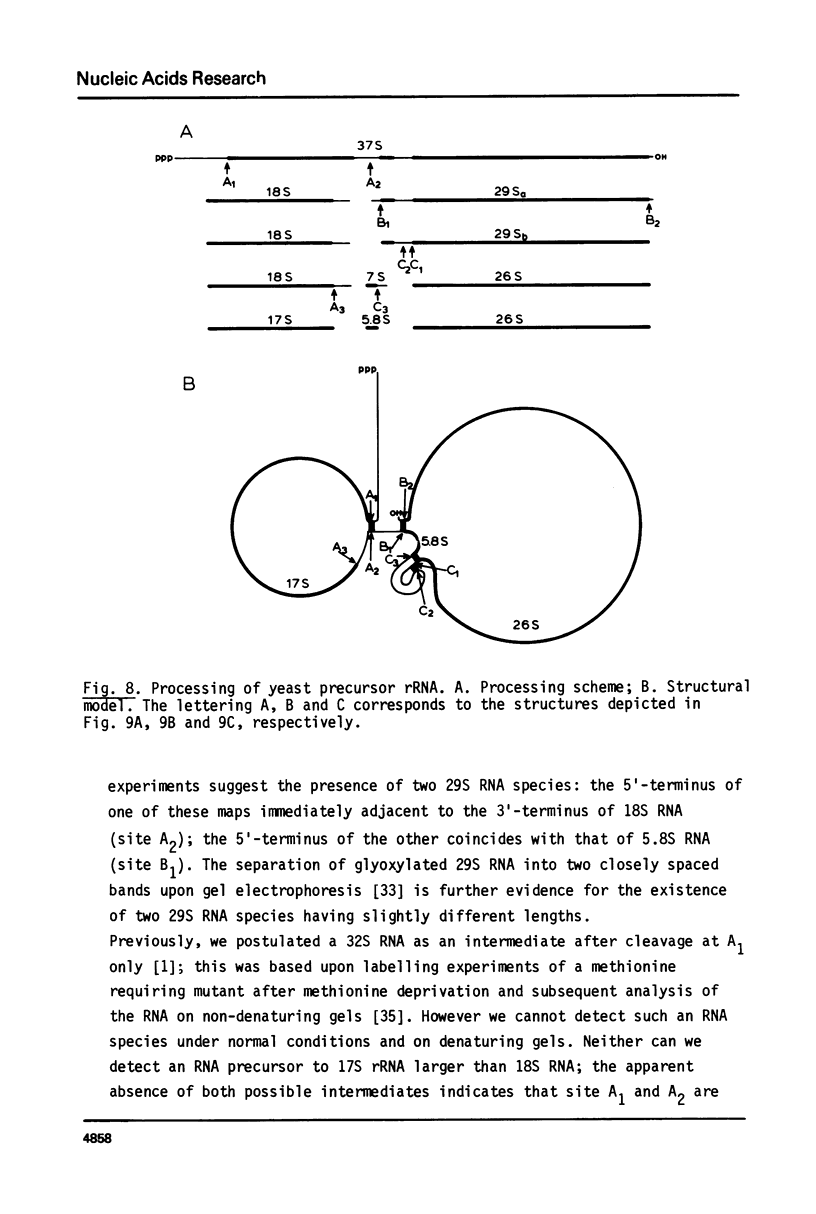
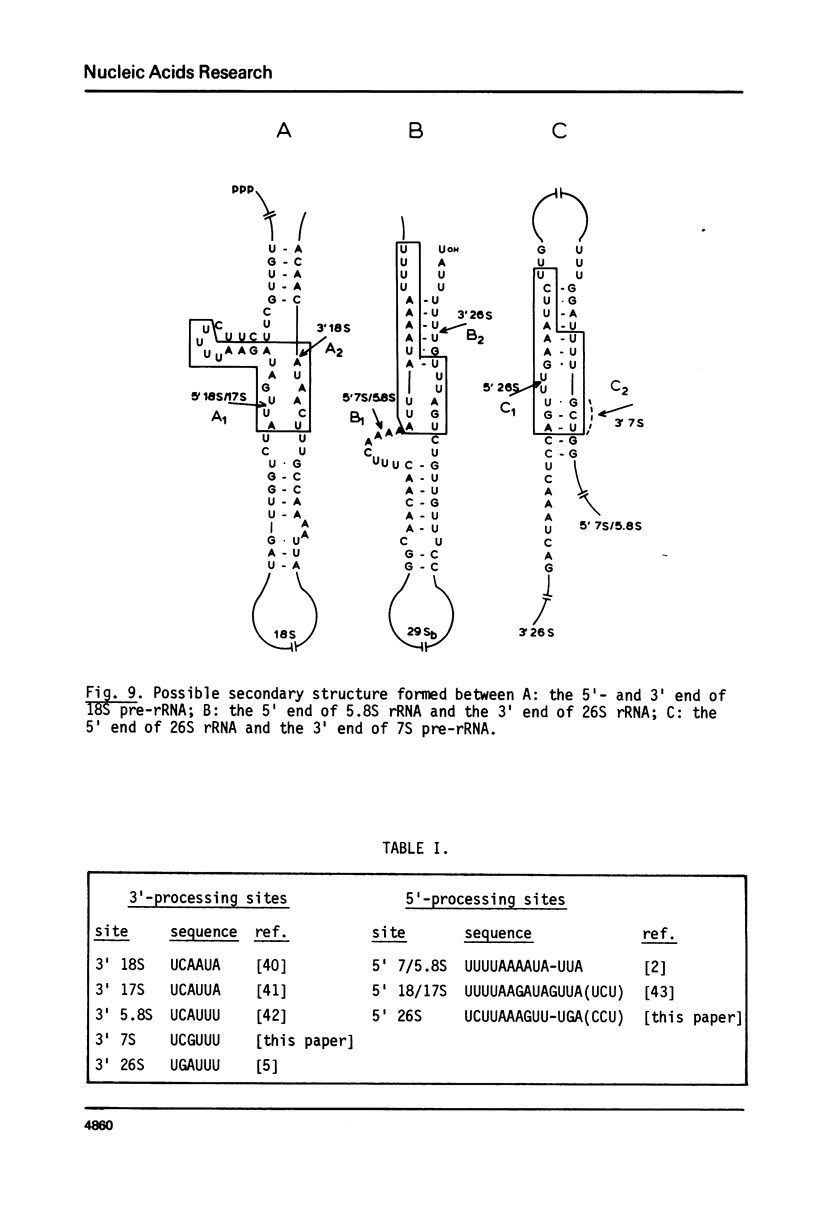
We have determined the nucleotide sequence of part of a cloned yeast ribosomal RNA operon extending from the 5.8S RNA gene downstream into the 5' -terminal region of the 26S RNA gene. We mapped the pertinent processing sites, viz. the 5' end of 26S rRNA and the 3'ends of 5.8S rRNA and its immediate precursor, 7S RNA. At the 3' end of 7S RNA we find the sequence UCGUUU which is very similar to the type I consensus sequence UCAUUA/U present at the 3' ends of 17S, 5.8S and 26S rRNA as well as 18S precursor rRNA in yeast. At the 5' end of the 26S RNA gene we find a sequence of thirteen nucleotides which is homologous to the type II sequence present at the 5' termini of both the 17S and the 5.8S RNA gene. These findings further support the suggestion put forward earlier (G.M. Veldman et al. (1980) Nucl. Acids Res. 8, 2907-2920) that both consensus sequences are involved in the recognition of precursor rRNA by the processing nuclease(s). We discuss a model for the processing of yeast rRNA in which a processing enzyme sequentially recognizes several combinations of a type I and a type II consensus sequence. We also describe the existence of a significant base complementarity between sequences in the 5' -terminal region of 26S rRNA and the 3' -terminal region of 5.8S rRNA. We suggest that base pairing between these sequences contributes to the binding between 5.8S and 26S rRNA.
Full text
PDF




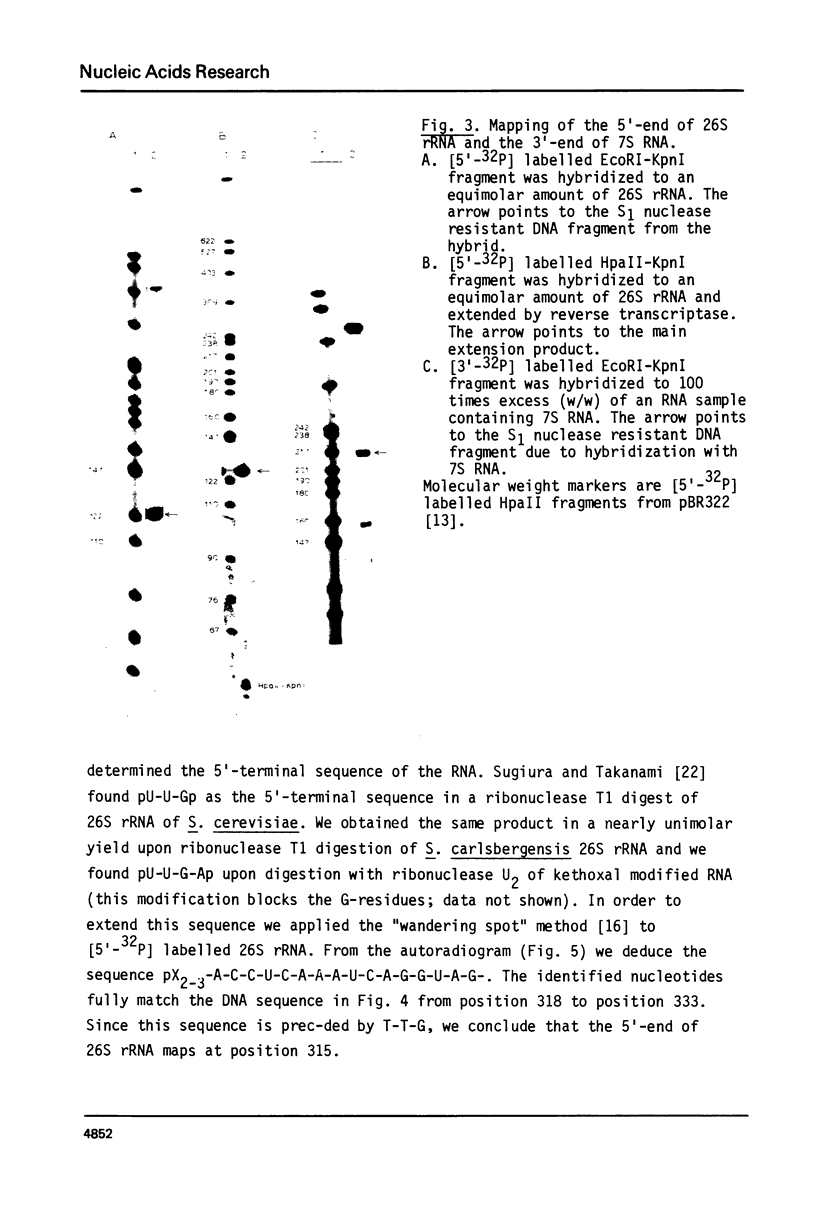
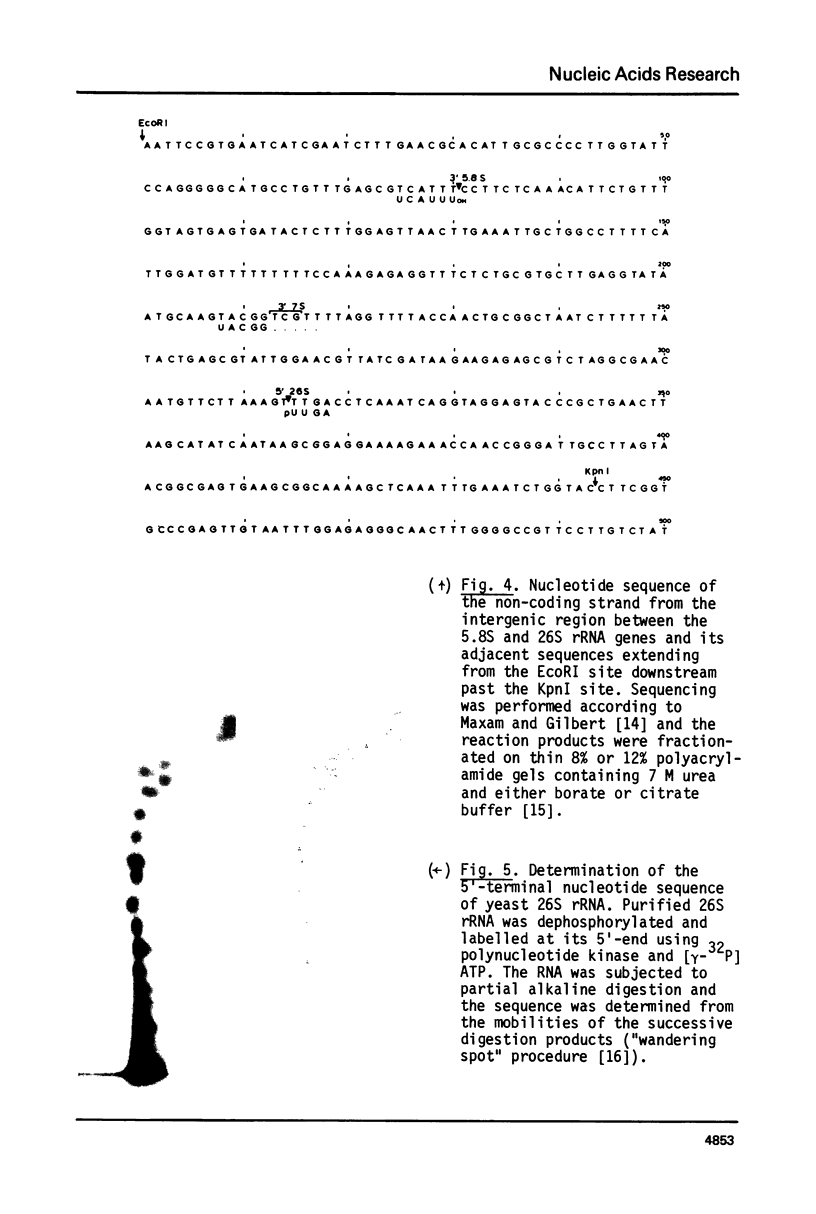






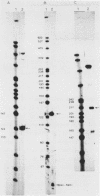
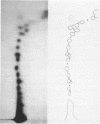
Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Abelson J. RNA processing and the intervening sequence problem. Annu Rev Biochem. 1979;48:1035–1069. doi: 10.1146/annurev.bi.48.070179.005131. [DOI] [PubMed] [Google Scholar]
- Bayev A., Georgiev O. I., Hadjiolov A. A., Nikolaev N., Skryabin K. G., Zakharyev V. M. The structure of the yeast ribosomal RNA genes. 3. Precise mapping of the 18 S and 25 S rRNA genes and structure of the adjacent regions. Nucleic Acids Res. 1981 Feb 25;9(4):789–799. doi: 10.1093/nar/9.4.789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bell G. I., DeGennaro L. J., Gelfand D. H., Bishop R. J., Valenzuela P., Rutter W. J. Ribosomal RNA genes of Saccharomyces cerevisiae. I. Physical map of the repeating unit and location of the regions coding for 5 S, 5.8 S, 18 S, and 25 S ribosomal RNAs. J Biol Chem. 1977 Nov 25;252(22):8118–8125. [PubMed] [Google Scholar]
- Berk A. J., Sharp P. A. Sizing and mapping of early adenovirus mRNAs by gel electrophoresis of S1 endonuclease-digested hybrids. Cell. 1977 Nov;12(3):721–732. doi: 10.1016/0092-8674(77)90272-0. [DOI] [PubMed] [Google Scholar]
- Boseley P. G., Tuyns A., Birnstiel M. L. Mapping of the Xenopus laevis 5.8S rDNA by restriction and DNA sequencing. Nucleic Acids Res. 1978 Apr;5(4):1121–1137. doi: 10.1093/nar/5.4.1121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bram R. J., Young R. A., Steitz J. A. The ribonuclease III site flanking 23S sequences in the 30S ribosomal precursor RNA of E. coli. Cell. 1980 Feb;19(2):393–401. doi: 10.1016/0092-8674(80)90513-9. [DOI] [PubMed] [Google Scholar]
- Brosius J., Dull T. J., Noller H. F. Complete nucleotide sequence of a 23S ribosomal RNA gene from Escherichia coli. Proc Natl Acad Sci U S A. 1980 Jan;77(1):201–204. doi: 10.1073/pnas.77.1.201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brownlee G. G., Sanger F. Chromatography of 32P-labelled oligonucleotides on thin layers of DEAE-cellulose. Eur J Biochem. 1969 Dec;11(2):395–399. doi: 10.1111/j.1432-1033.1969.tb00786.x. [DOI] [PubMed] [Google Scholar]
- De Jonge P., Kastelein R. A., Planta R. J. Non-ribosomal nucleotide sequences in 7-S RNA, the immediate precursor of 5.8-S ribosomal RNA in yeast. Eur J Biochem. 1978 Feb;83(2):537–546. doi: 10.1111/j.1432-1033.1978.tb12121.x. [DOI] [PubMed] [Google Scholar]
- De Jonge P., Klootwijk J., Planta R. J. Sequence of the 3'-terminal 21 nucleotides of yeast 17S ribosomal RNA. Nucleic Acids Res. 1977 Oct;4(10):3655–3663. doi: 10.1093/nar/4.10.3655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Donis-Keller H., Maxam A. M., Gilbert W. Mapping adenines, guanines, and pyrimidines in RNA. Nucleic Acids Res. 1977 Aug;4(8):2527–2538. doi: 10.1093/nar/4.8.2527. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goebel W., Bonewald R. Class of small multicopy plasmids originating from the mutant antibiotic resistance factor R1 drd-19B2. J Bacteriol. 1975 Aug;123(2):658–665. doi: 10.1128/jb.123.2.658-665.1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall L. M., Maden B. E. Nucleotide sequence through the 18S-28S intergene region of a vertebrate ribosomal transcription unit. Nucleic Acids Res. 1980 Dec 20;8(24):5993–6005. doi: 10.1093/nar/8.24.5993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jay E., Bambara R., Padmanabhan R., Wu R. DNA sequence analysis: a general, simple and rapid method for sequencing large oligodeoxyribonucleotide fragments by mapping. Nucleic Acids Res. 1974 Mar;1(3):331–353. doi: 10.1093/nar/1.3.331. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kelly J. M., Cox R. A. The nucleotide sequence at the 3'-end of Neurospora crassa 25S-rRNA and the location of a 5.8S-rRNA binding site. Nucleic Acids Res. 1981 Mar 11;9(5):1111–1121. doi: 10.1093/nar/9.5.1111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klemenz R., Geiduschek E. P. The 5' terminus of the precursor ribosomal RNA of Saccharomyces cerevisiae. Nucleic Acids Res. 1980 Jun 25;8(12):2679–2689. doi: 10.1093/nar/8.12.2679. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levis R., Dunsmuir P., Rubin G. M. Terminal repeats of the Drosophila transposable element copia: nucleotide sequence and genomic organization. Cell. 1980 Sep;21(2):581–588. doi: 10.1016/0092-8674(80)90496-1. [DOI] [PubMed] [Google Scholar]
- Loening U. E. The determination of the molecular weight of ribonucleic acid by polyacrylamide-gel electrophresis. The effects of changes in conformation. Biochem J. 1969 Jun;113(1):131–138. doi: 10.1042/bj1130131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maxam A. M., Gilbert W. A new method for sequencing DNA. Proc Natl Acad Sci U S A. 1977 Feb;74(2):560–564. doi: 10.1073/pnas.74.2.560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meyhack B., Pace B., Pace N. R. Involvement of precursor-specific segments in the in vitro maturation of Bacillus subtilis precursor 5S ribosomal RNA. Biochemistry. 1977 Nov 15;16(23):5009–5015. doi: 10.1021/bi00642a011. [DOI] [PubMed] [Google Scholar]
- Nazar R. N. A 5.8 S rRNA-like sequence in prokaryotic 23 S rRNA. FEBS Lett. 1980 Oct 6;119(2):212–214. doi: 10.1016/0014-5793(80)80254-7. [DOI] [PubMed] [Google Scholar]
- Nazar R. N., Sitz T. O. Role of the 5'-terminal sequence in the RNA binding site of yeast 5.8 S rRNA. FEBS Lett. 1980 Jun 16;115(1):71–76. doi: 10.1016/0014-5793(80)80729-0. [DOI] [PubMed] [Google Scholar]
- Pace N. R., Walker T. A., Schroeder E. Structure of the 5.8S RNA component of the 5.8S-28S ribosomal RNA junction complex. Biochemistry. 1977 Nov 29;16(24):5321–5328. doi: 10.1021/bi00643a025. [DOI] [PubMed] [Google Scholar]
- Pavlakis G. N., Jordan B. R., Wurst R. M., Vournakis J. N. Sequence and secondary structure of Drosophila melanogaster 5.8S and 2S rRNAs and of the processing site between them. Nucleic Acids Res. 1979 Dec 20;7(8):2213–2238. doi: 10.1093/nar/7.8.2213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philippsen P., Thomas M., Kramer R. A., Davis R. W. Unique arrangement of coding sequences for 5 S, 5.8 S, 18 S and 25 S ribosomal RNA in Saccharomyces cerevisiae as determined by R-loop and hybridization analysis. J Mol Biol. 1978 Aug 15;123(3):387–404. doi: 10.1016/0022-2836(78)90086-4. [DOI] [PubMed] [Google Scholar]
- Retèl J., Planta R. J. On the mechanism of the biosynthesis of ribosomal RNA in yeast. Biochim Biophys Acta. 1970 Dec 14;224(2):458–469. doi: 10.1016/0005-2787(70)90578-2. [DOI] [PubMed] [Google Scholar]
- Retèl J., Planta R. J. The investigation of the ribosomal RNA sites in yeast DNA by the hybridization technique. Biochim Biophys Acta. 1968 Dec 17;169(2):416–429. doi: 10.1016/0005-2787(68)90050-6. [DOI] [PubMed] [Google Scholar]
- Rubin G. M. The nucleotide sequence of Saccharomyces cerevisiae 5.8 S ribosomal ribonucleic acid. J Biol Chem. 1973 Jun 10;248(11):3860–3875. [PubMed] [Google Scholar]
- Sanger F., Coulson A. R. The use of thin acrylamide gels for DNA sequencing. FEBS Lett. 1978 Mar 1;87(1):107–110. doi: 10.1016/0014-5793(78)80145-8. [DOI] [PubMed] [Google Scholar]
- Skryabin K. G., Maxam A. M., Petes T. D., Hereford L. Location of the 5.8S rRNA gene of Saccharomyces cerevisiae. J Bacteriol. 1978 Apr;134(1):306–309. doi: 10.1128/jb.134.1.306-309.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugiura M., Takanami M. Analysis of the 5'-terminal nucleotide sequences of ribonucleic acids. II. Comparison of the 5'-terminal nucleotide sequences of ribosomal RNA's from different organisms. Proc Natl Acad Sci U S A. 1967 Oct;58(4):1595–1602. doi: 10.1073/pnas.58.4.1595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sutcliffe J. G. pBR322 restriction map derived from the DNA sequence: accurate DNA size markers up to 4361 nucleotide pairs long. Nucleic Acids Res. 1978 Aug;5(8):2721–2728. doi: 10.1093/nar/5.8.2721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiulenev V. I., Masiuk A. I. Nervnaia reguliatsiia sinteza RNK v iadrakh pecheni krys. Dokl Akad Nauk SSSR. 1979;247(5):1278–1280. [PubMed] [Google Scholar]
- Trapman J., Planta R. J. Maturation of ribosomes in yeast. I Kinetic analysis by labelling of high molecular weight rRNA species. Biochim Biophys Acta. 1976 Sep 6;442(3):265–274. doi: 10.1016/0005-2787(76)90301-4. [DOI] [PubMed] [Google Scholar]
- Trapman J., Retèl J., Planta R. J. Ribosomal precursor particles from yeast. Exp Cell Res. 1975 Jan;90(1):95–104. doi: 10.1016/0014-4827(75)90361-4. [DOI] [PubMed] [Google Scholar]
- Veldman G. M., Brand R. C., Klootwijk J., Planta R. Some characteristics of processing sites in ribosomal precursor RNA of yeast. Nucleic Acids Res. 1980 Jul 11;8(13):2907–2920. doi: 10.1093/nar/8.13.2907. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Veldman G. M., Klootwijk J., de Jonge P., Leer R. J., Planta R. J. The transcription termination site of the ribosomal RNA operon in yeast. Nucleic Acids Res. 1980 Nov 25;8(22):5179–5192. doi: 10.1093/nar/8.22.5179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker W. F. Proposed sequence homology between the 5'-end regions of prokaryotic 23 S rRNA and eukaryotic 28 S rRNA. Relevance to the hypothesis that 5.8 S rRNA is homologous to the 5'-end region of 23 S rRNA. FEBS Lett. 1981 Apr 20;126(2):150–151. doi: 10.1016/0014-5793(81)80228-1. [DOI] [PubMed] [Google Scholar]