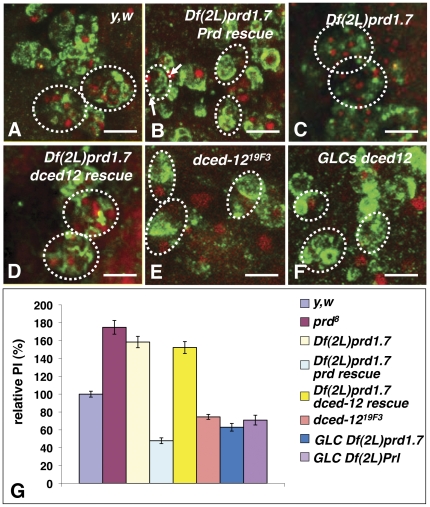
Figure 4. Dmel\ced-12 is required in macrophages for efficient clearance of apoptotic cells.
In A–F are projected confocal images of macrophages of stage 13 embryos stained with the anti-CRQ Ab (green) and where apoptotic corpses are stained with 7-AAD (red). In A, wild-type macrophages are shown that appear large as they engulf several apoptotic cells each. In B, prd-rescued Df(2L)prd1.7 homozygous mutant macrophages fail to efficiently engulf multiple corpses and appear small. Df(2L)prd1.7 homozygous mutant macrophages where a UAS-Dmel\ced-12transgene is being specifically over-expressed using a crq-gal4 driver on the third chromosome (D) do not engulf significantly more efficiently than in absence of the UAS-Dmel\ced-12transgene (C). In E and F are images of macrophages within the head of dced-12[19F3] homozygous mutant embryos (D) and that of macrophages of dced-12[PB] GLCs in trans- with Df(2L)prd1.7 (F). Macrophages in dced-12[19F3] and GLCs mutant embryos can still engulf apoptotic cells, albeit at a lower efficiency than that of wild-types (A). Scale bars in A–F are 5 µm. A graph in G summarizes the quantification of the PIs of wild-type (yw), Df(2L)prd1.7, prd-rescued Df(2L)prd1.7, dced-12-rescued Df(2L)prd1.7 on the third chromosome, dced-12[19F3], and Dmel\ced-12GLCs homozygous macrophages. Each bar represents the mean value ± SEM of the relative PIs for each genotype. Scale bars in panels A–F are 5 µm. In A–F, dotted white circles are indicative of individual macrophage cell bodies based on 7-AAD staining of their regular nuclei and CRQ staining.