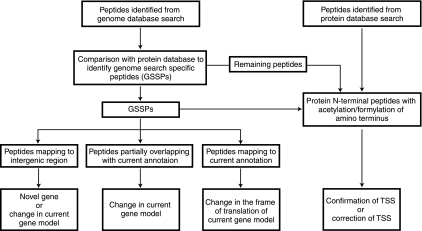
Fig. 3.
Schematic representation of proteogenomic annotation strategy. Peptide qualifying 1% FDR threshold from both protein and genome search were considered for proteogenomic analysis. From six frame translated genome database search results, peptides which mapped to known proteins from protein database were eliminated to obtain a set of genome search specific peptides (GSSPs). GSSPs were classified as mapping to intergenic region partially overlapping with current gene annotation and completely mapping within current gene annotations which were further used to modify the present genome annotation. N-terminal peptides identified from protein database searches were used to either confirm or propose change in the annotated translational start sites.