Abstract
Swi2/Snf2-type ATPases broadly regulate genome-associated processes such as transcription, replication and repair by catalyzing disruption, assembly, or remodeling of nucleosomes or other protein:DNA complexes1,2. ATP-driven motor activity along DNA has been suggested to disrupt target protein:DNA interactions in the remodeling reaction3–5. However, the complex and highly specific remodeling reactions are poorly understood, mostly because we lack high-resolution structural information on how remodelers bind their substrate proteins. Mot1 (modifier of transcription 1, denoted BTAF1 in humans) is a Swi2/Snf2 enzyme that specifically displaces TATA box binding protein (TBP) from promoter DNA and globally regulates transcription by generating a highly dynamic TBP pool in the cell6,7. As a Swi2/Snf2 enzyme that functions as a single polypeptide and interacts with a relatively simple substrate, Mot1 offers an ideal system for a better understanding of this important enzyme family. To reveal how Mot1 specifically disrupts TBP:DNA, we combined crystal and electron microscopy structures of Mot1:TBP complexes with biochemical studies. Here we show that Mot1 wraps around TBP and appears to act like a bottle opener: a spring-like array of 16 HEAT (huntingtin, elongation factor 3, PP2A and lipid kinase TOR) repeats grips the DNA distal side of TBP via loop insertions, while the Swi2/Snf2 domain binds upstream DNA, positioned to weaken TBPs DNA interaction by DNA translocation. A “latch” subsequently blocks TBP’s DNA binding groove, acting as a chaperone to prevent DNA re-association for efficient promoter clearance. This work shows how a remodeling enzyme can combine both motor and chaperone activities to achieve functional specificity using a conserved Swi2/Snf2 translocase.
Mot1 is highly conserved among eukaryotes and consists of an approx. 90–140 kDa N-terminal TBP binding region with predicted HEAT repeats followed by an approx. 60–70 kDa C-terminal Swi2/Snf2 type ATPase domain8,9. To provide a structural framework for a remodeler-substrate complex, we determined the crystal structure of Encephalitozoon cuniculi (Ec) Mot1 N-terminal domain (Mot1NTD; comprising the HEAT domain, residues 1–779, lacking the ATPase domain, residues 780–1256) in complex with full length EcTBP to 3.1 Å resolution (Fig. 1, Suppl. Table 1). EcMot1 possesses the characteristic sequence and biochemical features of Saccharomyces cerevisiae (Sc) Mot1 and human BTAF1, including TBP and DNA-stimulated ATPase activity, TBP binding via its HEAT domain, and – most importantly – ATP-stimulated TBP displacement from TATA DNA (Suppl. Figs. 1, 2).
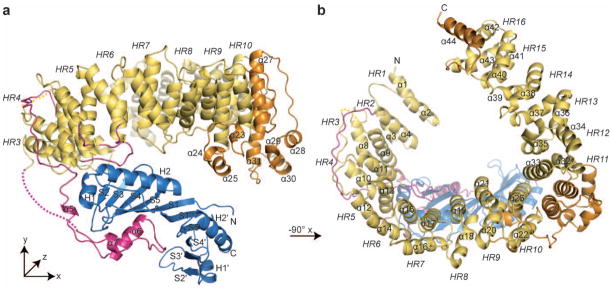
Figure 1. Overview of the EcMot1NTD EcTBP structure.
Panels (a) and (b) show the structure of the EcMot1NTD:EcTBP complex in ribbon representation with highlighted and annotated secondary structure. The HEAT repeats (HR) of EcMot1NTD are colored in yellow and non-HEAT repeat insertions in orange. The latch and the loops of HR 4 to HR 6 are highlighted in magenta. EcTBP is colored in blue. Two loops not traced by electron density are indicated by dashed lines.
The EcMot1NTD consists of a highly elongated stretch of 16 HEAT repeats (HRs), arranged in a horseshoe shape with approx. 95 Å x 85 Å x 40 Å dimensions and forms a specific 1:1 complex with EcTBP (Fig. 1). Remarkably, Mot1 wraps around one side of the pseudosymmetric TBP and grips both the convex protein interacting surface and the concave DNA binding surface of TBP via several loop insertions in the HR array. This wrapping interaction enables Mot1 to split the very stable EcTBP dimer that forms in the absence of DNA10 and that we observed biochemically and in a separate crystal structure of EcTBP alone at 1.9 Å resolution (Suppl. Fig. 3a, Suppl. Table 1). Despite this dual sided grip, Mot1 does not substantially alter the structure of TBP per se, since EcTBP bound to Mot1, EcTBP in the TBP dimer and, ScTBP bound to DNA are all very similar (Suppl. Fig. 3c–f). This suggests that remodeling of TBP does not proceed via changes in TBP structure as a simple consequence of Mot1 binding, but requires the ATP-dependent action of the Swi2/Snf2 domain.
Since promoter-bound TBP has its DNA binding surface occupied, Mot1 uses highly complementary HR loops to recognize the convex protein interaction surface of TBP (Fig. 2a). α-helices H1TBP and H2TBP are bound by the loop of HR 4 (residues 209–221) and by interactions with α13 in HR 5 and α15 in HR 6. The majority of these interactions are ion pairs between R46TBP, R48TBP, R65TBP, R96TBP, K99TBP, K103TBP and D212Mot1, D215Mot1, D216Mot1, Q256Mot1, D290Mot1 and D292Mot1 (Suppl. Table 2). In addition, F213Mot1 binds to a hydrophobic cleft between H1, H2 and β-sheet S2 and provides a hydrophobic anchor, while F210Mot1 and W255Mot1 pack against the side chains of R48TBP and K103TBP.
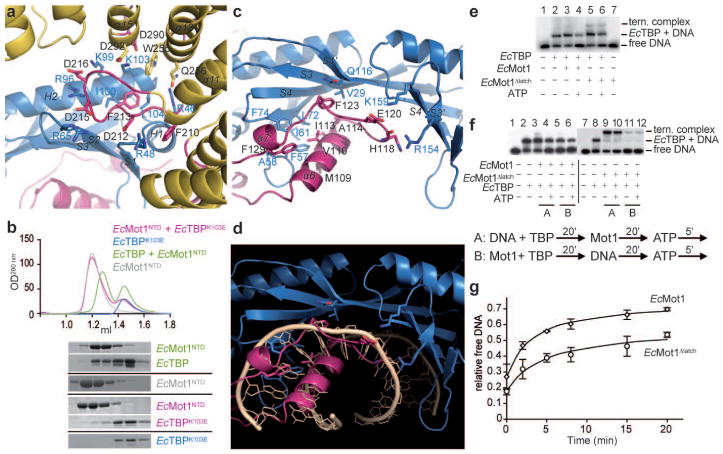
Figure 2. Details of the interaction interfaces and latch function.
a, Close-up view of the EcMot1:EcTBP interaction (color scheme of Fig. 1) b, Wildtype EcTBP and EcMot1NTD (green) can form a stable complex, whereas EcTBPK103E mutant doesn’t coelute with EcMot1NTD (pink) in size exclusion chromatography (Suppl. Fig. 1b). EcMot1’s latch (pink, shown in panel c) overlaps with the DNA-binding region (shown in panel d) of EcTBP (blue). Some bases of the superimposed DNA (wheat, from pdb 1YTB29) were omitted. e, f, EMSAs (for corresponding quantitations see Suppl. Fig.4). EcMot1 Δlatch formed stable ternary complexes with EcTBP:DNA (lane 5). However, while wildtype EcMot1 largely cleared the DNA probe of bound TBP in an ATP-dependent reaction (lane 4), EcMot1 Δlatch was less efficient in TBP removal (lane 6). f, EcMot1 was incubated with EcTBP after (A) or prior to the addition of DNA (B). Preincubation of the two proteins inhibits TBP’s ability to bind DNA. g, EcMot1 Δlatch was capable of less efficient EcTBP:DNA dissociation than wild-type EcMot1. ATP was added to preformed EcMot1:EcTBP:DNA or EcMot1 Δlatch:EcTBP:DNA ternary complexes, and the proportion of free DNA was quantified by EMSA at various times thereafter. Data represent mean and standard error from two independent experiments.
These interactions are evolutionary well conserved (Suppl. Fig. 4a; Suppl. Table 2) and supported by functional data in vivo and in vitro. For instance, K145ScTBP (K103EcTBP) is an essential residue for stabilization of the ScMot1:ScTBP interaction8. We mutated K103 in EcTBP and observed that EcTBPK103E failed to form a stable complex with EcMot1NTD in vitro (Fig. 2b). Moreover, mutation in D365 (D212EcMot1) inactivated ScMot1 function in vivo and abolished the Mot1:TBP interaction in vitro8. Mutations in ScTBP K138 also impaired the interaction with ScMot1, consistent with the projection of the homologous side chains into the EcMot1NTD:EcTBP interface8,11. The distribution of residues along the length of the EcMot1NTD is also consistent with prior work showing that broad segments of the ScMot1 and BTAF1 N-terminus are important for stable interaction with TBP8,9,12. Thus, the specific interaction interface between the Mot1 HRs and the convex surface of TBP is well suited to provide specific recognition of the TBP surface in the TBP:promoter complex, explaining why Mot1 specifically targets TBP:DNA and not other protein:DNA complexes.
Unexpectedly, TBP’s concave DNA binding surface, obviously accessible only when TBP is displaced from promoter DNA, is bound by Mot1 as well (Fig. 2c). A long “latch” located between HRs 2 and 3, protrudes from the side of Mot1 distal to TBP, and wraps all the way around the side of Mot1 and TBP. Remarkably, its tip (residues 101–130) substitutes for interactions made by four base pairs at and immediately downstream from the TATA sequence (Fig. 2d). A set of hydrophobic interactions matches the hydrophobic nature of TBP’s DNA binding groove. For instance, the side chain of M109Mot1 replaces a deoxyribose moiety in binding to F57TBP, a prominent and highly conserved DNA binding residue of TBP. The main chain of residues 118–129 folds along the position of the backbone of the coding DNA strand, with side chains often placed at positions occupied by base and sugar moieties. F123Mot1 replaces a deoxyribose moiety and stacks with the conserved Q116TBP, while F129Mot1 replaces a base moiety that interacts with the aromatic pair F57TBP and F74TBP.
To test the function of the latch, we generated EcMot1 Δlatch and EcMot1NTD Δlatch mutants that lack residues 96–132. Both proteins can still interact with EcTBP with approx. equal Mot1:TBP molarity (Suppl. Fig. 4b). This observation suggests that EcTBP is mainly bound by acidic loops of Mot1’s HR 4–6. However, the latch might prevent TBP rebinding to DNA (after DNA dissociation) and homodimerization by saturating the exposed, hydrophobic DNA binding cleft of TBP (see Suppl. Fig. 3b). Indeed, whereas EcMot1NTD forms a heterodimer with EcTBP, we find that EcMot1NTD Δlatch forms a 2:2 complex with EcTBP (Suppl. Table 3). The most likely explanation is that two EcMot1NTD Δlatch molecules bind the EcTBP dimer, but fail to dissociate the TBP dimer due to the absence of the latch. Since EcMot1 Δlatch in complex with EcTBP doesn’t show a substantially increased hydrodynamic radius compared to the WT complex in gel filtration (Suppl. Fig 4b), it is likely that the Swi2/Snf2 domain sterically prevents dimerization of EcMot1 Δlatch via TBP dimers.
Thus, while one function of the latch might be to keep TBP in a monomeric state, a perhaps more intriguing role of the latch is to interfere with DNA binding by TBP. To test this, we analyzed the ability of the EcMot1 Δlatch protein to bind to the TBP:DNA complex. In contrast to wild-type EcMot1, EcMot1 Δlatch formed readily detectable ternary complexes with EcTBP and DNA (Fig. 2e, f), indicating that the latch makes the association of EcMot1 with EcTBP:DNA less stable. Although it binds TBP:DNA more efficiently, EcMot1 Δlatch was notably impaired in ATP-dependent TBP:DNA dissociation (Figure 2e–g, Suppl. Fig. 4d, e). This was not due to a defect in the ATPase activity (Suppl. Fig. 4g). Moreover, when combined with EcTBP prior to DNA addition, EcMot1 inhibited DNA binding by EcTBP (Fig. 2f, Suppl. Fig. 4e). EcMot1NTD also inhibited DNA binding by EcTBP in a reaction that required the latch (Suppl. Fig. 4c, f). However, the latch was not essential for inhibiting the EcTBP:DNA interaction in the context of the full-length EcMot1 protein (Fig. 2f, Suppl. Fig. 4e), indicating that both the latch and ATPase domains can modulate EcTBP DNA binding activity. Taken together, the data suggest that the latch has “chaperone” activity that regulates macromolecular interactions with TBP’s hydrophobic groove. Since DNA and latch binding to TBP are mutually exclusive (Fig. 2d), it is unlikely that the latch initially disrupts the TBP:DNA complex. Consistent with this, EcMot1 Δlatch was able to displace TBP from DNA using ATP, but the overall level of displacement was increased by the latch (Fig. 2g). Thus, our combined data can be explained by a physiologically plausible model where the ATP-dependent action of the Swi2/Snf2 domain remodels TBP:TATA first, and then the latch blocks the exposed hydrophobic groove to prevent rebinding.
To reveal the architecture of the whole E. cuniculi Mot1:TBP complex including its Swi2/Snf2 domain, we generated 3D reconstructions of negatively stained EcMot1:EcTBP particles visualized in electron micrographs (Fig. 3a, Suppl. Fig. 5). The 3D reconstruction is shaped like a slightly closed “C” with a globular protrusion and is similar to the 3D reconstructions of the human TBP:BTAF1 complex13. To unambiguously locate the Swi2/Snf2 domain, an EcMot1ΔCT:EcTBP complex was imaged, in which the C-terminal half of the Swi2/Snf2 domain was truncated (Suppl. Figs. 5, 6c). We find that the prominent protrusion is missing from this complex, suggesting that this protrusion corresponds to the C-terminal half of the ATPase (Fig. 3b). Finally, we imaged Mot1 without TBP (Suppl. Figs. 5, 6b). Although Mot1 alone is evidently more flexible and adopts a slightly different conformation than in the Mot1:TBP complex, not unexpected for a large HEAT array, a particular lateral density patch was seen to be missing, thereby defining the location of TBP in the complex. Altogether, these data allowed us to convincingly rigid body dock the Mot1NTD:TBP crystal structure into the EM density (Suppl. Fig. 6a).
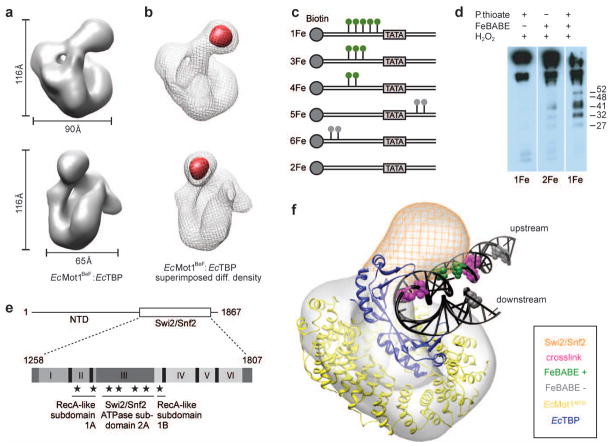
Figure 3. 3D reconstruction of the EcMot1:EcTBP complex and model of the EcMot1:EcTBP:DNA complex.
a, Two views of the EcMot1BeF:EcTBP density. ADP-BeF3− was added due to assumed stabilization of the ATPase domain b, Subtraction map (red) between EcMot1BeF:EcTBP (grey mesh) and EcMot1 ΔCT:EcTBP density maps. c, DNA probes with phosphorothioates (green/grey lollipops) used in FeBABE cleavage assays. d, FeBABE-mediated cleavage of Mot1 analyzed by Western blot30 with approximate sizes of the cleavage products in kDa. e, Summary of FeBABE results. Stars represent approximate sites of cleavage mediated by FeBABE conjugated to the DNA upstream of the TATA Box. f, Model of the Mot1:TBP:DNA complex. Electron density map of EcMot1BeF:EcTBP complexes with the crystal structure of EcMot1NTD:EcTBP including a superimposed elongated DNA from ScTBP:DNA complex (1YTB). Bases that represent 5-I dU substitutions used for crosslinking ScMot1 to DNA15 and bases that represent FeBABE probe 4Fe (Suppl. Fig. 7a) are colored in magenta and green, respectively. Positions of FeBABE conjugation that did not produce cleavage are colored in gray. The position of the Swi/Snf2 domain of Mot1 is indicated as an orange mesh.
To corroborate this placement, we superimposed TBP in the crystal structure with the ScTBP:DNA complex and extended the ends of the DNA with generic B-form DNA. Indeed, the “upstream” DNA protrudes toward the electron density corresponding to the Swi2/Snf2 domain in the EM 3D reconstruction (Fig. 3f). Our model predicts that the Swi2/Snf2 domain contacts the DNA around 10–17 bases upstream from the TATA sequence, well positioned to translocate along the minor groove of the DNA3. This is in good agreement with previous crosslinking results and satisfactorily explains why a duplex DNA extension is required upstream of the TBP binding site for formation of a catalytically active yeast Mot1:TBP:DNA complex14,15. To further validate this model, we localized the region of yeast Mot1 proximal to the upstream DNA using FeBABE-mediated hydroxyl radical cleavage16 (Suppl. Fig. 7a and Fig. 3c). As predicted by the model, FeBABE molecules positioned within a 9 bp DNA segment immediately upstream of the TATA sequence generated multiple specific C-terminal Mot1 fragments (cleavage in the Swi2/Snf2 domain), while no cleavage products were detected without FeBABE or when FeBABE molecules were conjugated to DNA upstream of this region or downstream of the TATA sequence (Fig. 3d,e and Suppl. Fig. 7b).
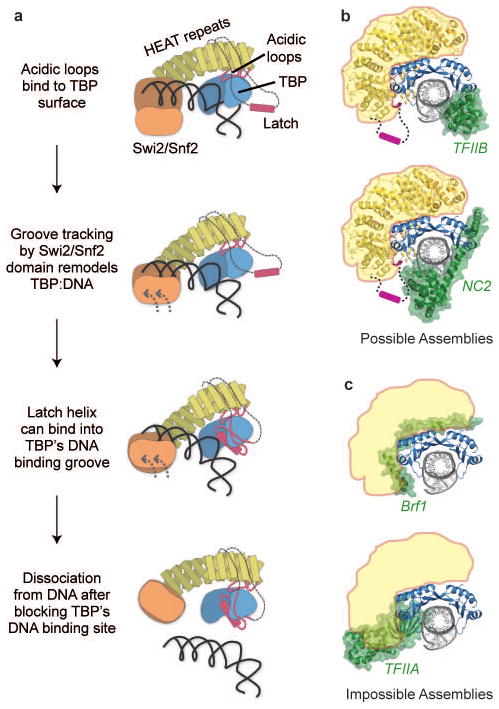
Our combined data suggest that Mot1 recognizes TATA-bound TBP by binding to the positively charged TBP surface at H1 and H2, and by binding of the Swi2/Snf2 domain to the minor groove of upstream DNA. We suggest that ATP-dependent groove tracking of the Swi2/Snf2 domain initially disrupts TBP:TATA, followed by binding of the latch to the exposed hydrophobic groove of TBP and full dissociation of Mot1:TBP from DNA (Fig. 4a). In this model, consistent with the translocation direction inferred for nucleosome remodeling enzymes17, the Swi2/Snf2 domain “pulls” on TBP. Alternatively, the Swi2/Snf2 domain might “push” TBP. The precise tracking directionality must await future studies, although the proposed two-step displacement could occur by translocation in either direction. In any case, the rotational force generate by even tracking a few base-pairs of DNA by the Swi2/Snf2 domain could sufficiently lift TBP from DNA such that the latch can bind. The energy of a few ATP-dependent translocation steps could be stored elastically in the HEAT repeats. In this way Mot1 would act like a bottle opener to lift TBP from DNA, with the acidic loops functioning as the head, the HRs as the handle, and the Swi2/Snf2 domain as the twisting hand.
Figure 4. Proposed remodeling mechanism.
a, Proposed mechanism of Mot1 mediated displacement of TBP from the DNA. b,c, Models of possible Mot1 substrates generated by superimposing the EcMot1:EcTBP crystal structure on other TBP containing structures. b, Possible Mot1 (yellow) substrates are TBP complexes with TFIIB (pdb code 1AIS) and NC2 (pdb code 1JFI). The Mot1 latch is omitted from the structure but drawn as a cartoon. c, Sterically impossible Mot1 (indicated in yellow) substrates are TBP complexes with TFIIA (pdb code 1NH2) or TFIIIB subunit Brf1 (pdb code 1NGM).
Since TBP exists in many different complexes that could be substrates for Mot1’s remodeling activity, we compared the Mot1:TBP complex with other structurally characterized TBP complexes. The HEAT domain of Mot1 would be able to interact with TBP:TFIIB:DNA complexes as well as with TBP:NC2:DNA complexes (Fig. 4b). The compatibility of Mot1 and NC2 binding to TBP-DNA is consistent with numerous in vitro and in vivo results8,18–21, including recent genome wide chromatin co-localization of Mot1 and NC220. In contrast, Mot1 sterically overlaps with TFIIA, explaining how Mot1 and TFIIA compete for binding to TBP (Fig. 4c)10,11,22. In addition, Mot1 evidently also clashes with Brf1, a subunit of the Pol III initiation factor TFIIIB (Fig. 4c), while we do not see any clashes with a recent TBP-TFIIB-Pol II preinitiation complex (PIC) model (Suppl. Fig. 8)23,24. Thus, these comparisons suggest that Mot1 can act on specific subsets of PICs in addition to TBP alone. These may include minimal PICs and incomplete PICs as well as NC2-repressed TBP complexes, while perhaps PICs, which include TFIIA and TAFs, or Pol III PICs (containing Brf1), may be excluded from regulation by Mot1.
The discovery of the latch and its role in reducing DNA binding and TBP dimerization indicates that Mot1 evidently not only displaces TBP, but blocks its hydrophobic surface patch to prevent interactions with DNA or other factors that bind to the concave surface. This activity argues that Mot1 acts as a TBP “chaperone” to control its interaction with other macromolecules. Mot1 might hold TBP in a diffusible state, explaining how it helps to rapidly redistribute TBP between different promoters and binding sites in the genome. Redistribution between promoters requires large diffusion steps between chromosomes and chromosome loops in trans, as opposed to sliding along DNA in cis, which is likely part of the repression mode of NC225. This model is supported by the important role of Mot1 on the high cellular mobility of TBP6,7 and by early findings that substantial proportions of TBP reside in a stable complex with Mot1 in HeLa and yeast cell extracts26,27.
The necessity of the peculiar Mot1:TBP interactions might come from the high affinity hydrophobic DNA binding mode of TBP as well as the necessity to tightly regulate its binding to specific sites in the genome, while preventing non-specific DNA interactions. Thus, a combination of motor and chaperone functions could be a more general feature of remodeling systems that deal with the assembly or disassembly of sticky proteins from DNA. In other systems, remodeling and chaperone functions may be provided by separate factors, as seen for example in the cooperation of the SWI/SNF nucleosome remodeling complex and Asf1p histone chaperone28.
In summary, the results here provide a high-resolution view of how a Swi2/Snf2-type remodeler interacts with its substrate, show how the conserved ATP-dependent DNA translocase module can be used to generate the high functional specificities within the large and diverse family of Swi2/Snf2 enzymes, and provide a testable mechanism for a remodeling reaction.
Methods Summary
Recombinant proteins coding for full-length EcMot1 (residues 1–1275), EcTBP, EcMot1 Δ CT (residues 1–1016), EcMot1NTD (residues 1–779) and EcMot1 Δlatch (Δ96–132) were produced in E. coli or in insect cells. Protein purification was conducted using standard methods and proteins were crystallized by hanging drop vapour diffusion. EcTBP crystals diffracted to 1.9 Å resolution and were measured at the Swiss Light Source (SLS, Villigen). Native data of crystals from EcMot1NTD:EcTBP diffracted X-rays to 3.1 Å, and were collected at the European Synchrotron Radiation Facility (ESRF, Grenoble). Derivative crystals of selenomethionine labelled EcMot1NTD:EcTBP were collected to 3.3 Å at the SLS. The structure of EcTBP was solved by molecular replacement using yeast TBP (1TBP) as a search model. The structure of EcMot1NTD:EcTBP was determined using selenium single-wavelength anomalous dispersion in combination with molecular replacement with the EcTBP structure as a partial model. EcMot1:EcTBP in presence of 2mM ADP and beryllium fluoride (ADP-BeF3−), EcMot1E912Q (Walker B mutant of EcMot1 instead of wildtype was used due to enhanced stability) or EcMot1 ΔCT:EcTBP were used for negative stain (2 % uranyl acetate) electron microscopic studies and micrographs were recorded on a Tecnai G2 Spirit TEM at 120 kV. Size exclusion experiments were performed on Ettan LC system (GE Healthcare, Superose 12 PC 3.2/30). FeBABE (Dojindo) was conjugated to 68 bp DNA duplexes, based on the sequence of the adenovirus major late promoter. Biotinylation of the top strand’s 5′ end allowed the duplexes to be bound by streptavidin beads. Following FeBABE conjugation, TBP and Mot1 were loaded onto the modified DNAs and cutting was initiated by addition of ascorbic acid and hydrogen peroxide.
Full Methods and any associated references are available in the online version of the paper at www.nature.com/nature.
Supplementary Material
Acknowledgments
We thank the Max-Planck Crystallization Facility Martinsried. We thank Maria Lucas, Alexandra Schele, Charlotte Ungewickell, Julia Goetzl and Yoshitaka Hiruma for help with experimentation. We thank Jean-Paul Armache and Martin Turk for help with EM data. We are grateful to Gail Miller and Steve Hahn for advice. We thank the staffs at the SLS (Villingen) and ESRF (Grenoble) for help with data collection. We thank Patrick Cramer and members of the Hopfner and Auble labs for discussions and comments on the manuscript. This work was supported by the German Research Council (SFB 646 and SFB/TR5) and Excellence Initiative (Center for Integrated Protein Science Munich) to K.-P.H. and R.B., by DFG grant WE4628/1 to P.We. and by NIH grant GM55763 to D.T.A.
Footnotes
Suppl. Information is linked to the online version of the paper at www.nature.com/nature.
Author Contributions S.C. and M.M. cloned, purified, crystallized 3OIC and S.C. solved its structure. S.C., A.B., M.M. and P.W. cloned, purified, crystallized 3OC3 and P.W. collected data and P.W., G.W. and K.P.H. solved the complex structures. R.V. performed FeBABE experiments, M.N.W. conducted yeast molecular biological manipulations and D.T.A. performed gelshifts. P.We., O.B. and R.B. performed and interpreted EM data. P.W., P.We, R.B., D.T.A. and K.-P.H. planned and interpreted the experiments. D.T.A. and K.-P.H. wrote manuscript and all authors provided editorial input.
Atomic coordinates and structure factors for the reported crystal structures have been deposited with the Protein Data Bank under accession codes 3OCI (EcTBP) and 3OC3 (EcTBP:EcMot1NTDcomplex).
Reprints and permissions information is available at www.nature.com/reprints. The authors declare no competing financial interests: details accompany the full-text HTML version of the paper at www.nature.com/nature.
References
- 1.Cairns BR. The logic of chromatin architecture and remodelling at promoters. Nature. 2009;461:193–198. doi: 10.1038/nature08450. [DOI] [PubMed] [Google Scholar]
- 2.Li B, Carey M, Workman JL. The role of chromatin during transcription. Cell. 2007;128:707–719. doi: 10.1016/j.cell.2007.01.015. [DOI] [PubMed] [Google Scholar]
- 3.Durr H, Korner C, Muller M, Hickmann V, Hopfner KP. X-ray structures of the Sulfolobus solfataricus SWI2/SNF2 ATPase core and its complex with DNA. Cell. 2005;121:363–373. doi: 10.1016/j.cell.2005.03.026. [DOI] [PubMed] [Google Scholar]
- 4.Saha A, Wittmeyer J, Cairns BR. Chromatin remodeling by RSC involves ATP-dependent DNA translocation. Genes Dev. 2002;16:2120–2134. doi: 10.1101/gad.995002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Racki LR, et al. The chromatin remodeller ACF acts as a dimeric motor to space nucleosomes. Nature. 2009;462:1016–1021. doi: 10.1038/nature08621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Auble DT. The dynamic personality of TATA-binding protein. Trends Biochem Sci. 2009;34:49–52. doi: 10.1016/j.tibs.2008.10.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.de Graaf P, et al. Chromatin interaction of TATA-binding protein is dynamically regulated in human cells. Journal of cell science. 2010;123:2663–2671. doi: 10.1242/jcs.064097. [DOI] [PubMed] [Google Scholar]
- 8.Darst RP, et al. Mot1 regulates the DNA binding activity of free TATA-binding protein in an ATP-dependent manner. The Journal of biological chemistry. 2003;278:13216–13226. doi: 10.1074/jbc.M211445200. [DOI] [PubMed] [Google Scholar]
- 9.Pereira LA, van der Knaap JA, van den Boom V, van den Heuvel FA, Timmers HT. TAF(II)170 interacts with the concave surface of TATA-binding protein to inhibit its DNA binding activity. Molecular and cellular biology. 2001;21:7523–7534. doi: 10.1128/MCB.21.21.7523-7534.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pugh BF. Control of gene expression through regulation of the TATA-binding protein. Gene. 2000;255:1–14. doi: 10.1016/s0378-1119(00)00288-2. [DOI] [PubMed] [Google Scholar]
- 11.Auble DT, Hahn S. An ATP-dependent inhibitor of TBP binding to DNA. Genes & development. 1993;7:844–856. doi: 10.1101/gad.7.5.844. [DOI] [PubMed] [Google Scholar]
- 12.Mohibullah N, Hahn S. Site-specific cross-linking of TBP in vivo and in vitro reveals a direct functional interaction with the SAGA subunit Spt3. Genes & development. 2008;22:2994–3006. doi: 10.1101/gad.1724408. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pereira LA, et al. Molecular architecture of the basal transcription factor B-TFIID. The Journal of biological chemistry. 2004;279:21802–21807. doi: 10.1074/jbc.M313519200. [DOI] [PubMed] [Google Scholar]
- 14.Darst RP, Wang D, Auble DT. MOT1-catalyzed TBP-DNA disruption: uncoupling DNA conformational change and role of upstream DNA. Embo J. 2001;20:2028–2040. doi: 10.1093/emboj/20.8.2028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sprouse RO, Brenowitz M, Auble DT. Snf2/Swi2-related ATPase Mot1 drives displacement of TATA-binding protein by gripping DNA. Embo J. 2006;25:1492–1504. doi: 10.1038/sj.emboj.7601050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Miller G, Hahn S. A DNA-tethered cleavage probe reveals the path for promoter DNA in the yeast preinitiation complex. Nature structural & molecular biology. 2006;13:603–610. doi: 10.1038/nsmb1117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gangaraju VK, Prasad P, Srour A, Kagalwala MN, Bartholomew B. Conformational changes associated with template commitment in ATP-dependent chromatin remodeling by ISW2. Mol Cell. 2009;35:58–69. doi: 10.1016/j.molcel.2009.05.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Dasgupta A, Darst RP, Martin KJ, Afshari CA, Auble DT. Mot1 activates and represses transcription by direct, ATPase-dependent mechanisms. Proceedings of the National Academy of Sciences of the United States of America. 2002;99:2666–2671. doi: 10.1073/pnas.052397899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Klejman MP, et al. NC2alpha interacts with BTAF1 and stimulates its ATP-dependent association with TATA-binding protein. Molecular and cellular biology. 2004;24:10072–10082. doi: 10.1128/MCB.24.22.10072-10082.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van Werven FJ, et al. Cooperative action of NC2 and Mot1p to regulate TATA-binding protein function across the genome. Genes & development. 2008;22:2359–2369. doi: 10.1101/gad.1682308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hsu JY, et al. TBP, Mot1, and NC2 establish a regulatory circuit that controls DPE-dependent versus TATA-dependent transcription. Genes & development. 2008;22:2353–2358. doi: 10.1101/gad.1681808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Geisberg JV, Struhl K. Cellular stress alters the transcriptional properties of promoter-bound Mot1-TBP complexes. Molecular cell. 2004;14:479–489. doi: 10.1016/j.molcel.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 23.Kostrewa D, et al. RNA polymerase II-TFIIB structure and mechanism of transcription initiation. Nature. 2009;462:323–330. doi: 10.1038/nature08548. [DOI] [PubMed] [Google Scholar]
- 24.Liu X, Bushnell DA, Wang D, Calero G, Kornberg RD. Structure of an RNA polymerase II-TFIIB complex and the transcription initiation mechanism. Science. 327:206–209. doi: 10.1126/science.1182015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Schluesche P, Stelzer G, Piaia E, Lamb DC, Meisterernst M. NC2 mobilizes TBP on core promoter TATA boxes. Nat Struct Mol Biol. 2007;14:1196–1201. doi: 10.1038/nsmb1328. [DOI] [PubMed] [Google Scholar]
- 26.Timmers HT, Meyers RE, Sharp PA. Composition of transcription factor B–TFIID. Proc Natl Acad Sci U S A. 1992;89:8140–8144. doi: 10.1073/pnas.89.17.8140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Poon D, Campbell AM, Bai Y, Weil PA. Yeast Taf170 is encoded by MOT1 and exists in a TATA box-binding protein (TBP)-TBP-associated factor complex distinct from transcription factor IID. The Journal of biological chemistry. 1994;269:23135–23140. [PubMed] [Google Scholar]
- 28.Gkikopoulos T, Havas KM, Dewar H, Owen-Hughes T. SWI/SNF and Asf1p cooperate to displace histones during induction of the saccharomyces cerevisiae HO promoter. Mol Cell Biol. 2009;29:4057–4066. doi: 10.1128/MCB.00400-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kim Y, Geiger JH, Hahn S, Sigler PB. Crystal structure of a yeast TBP/TATA-box complex. Nature. 1993;365:512–520. doi: 10.1038/365512a0. [DOI] [PubMed] [Google Scholar]
- 30.Auble DT, Wang D, Post KW, Hahn S. Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA. Molecular and cellular biology. 1997;17:4842–4851. doi: 10.1128/mcb.17.8.4842. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.