Fig. 2.
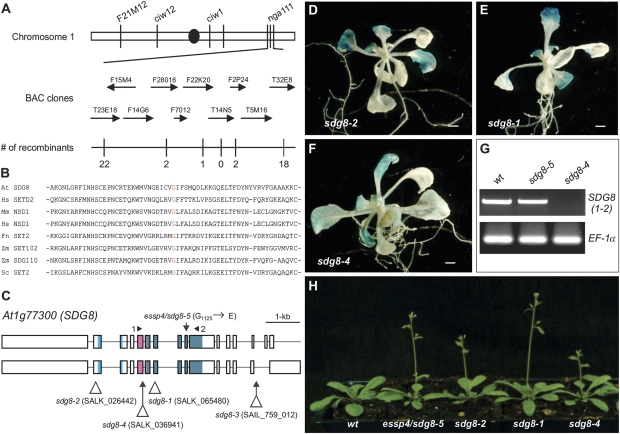
Map-based cloning of essp4. (A) Fine genetic mapping with PCR-based markers located the essp4 locus to the bottom of chromosome 1, on BAC clone T14N5. The numbers of recombination events out of the total numbers of chromosomes examined (1536) are indicated. (B) Alignment of amino acid sequences of SET domains from Arabidopsis (At), human (Hs), mouse (Mm), fungus (Fn), maize (Zm), and yeast (Sc). (C) Structure of the SDG8/ESSP4 gene and the location of mutation/T-DNA insertion sites of sdg8 alleles. Boxes and lines represent exons and introns, respectively. The shaded boxes represent the conserved protein domains (from left to right): CW (cysteine and tryptophan conserved), AWS (associated with SET), and SET. (D–F) GUS phenotypes of three T-DNA insertion alleles. Shown here is a representative F2 progeny from each of the crosses of the corresponding T-DNA allele with the βCGpro:GUS line. (G) RT-PCR analysis of the expression of SDG8 in the wild type and sdg8 mutants. The primers used are indicated in (C) and elongation factor 1α was used as an internal control. (H) Comparison of sdg8 mutant plants with the wild type at bolting. (This figure is available in colour at JXB online.)