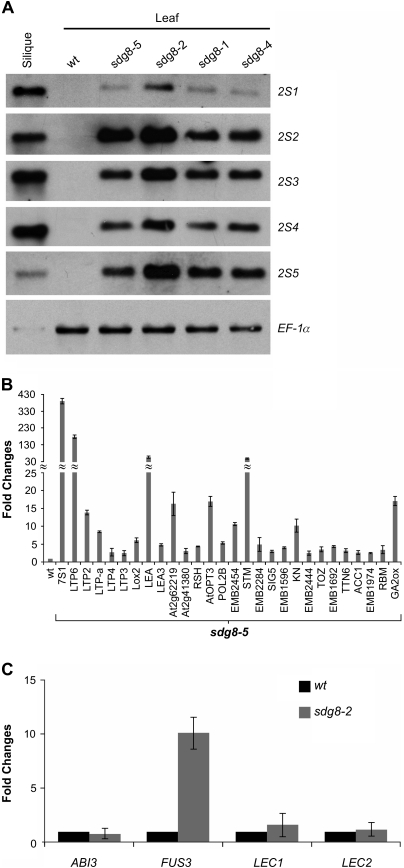
Fig. 3.
Expression analysis of seed maturation genes in essp4 mutant leaves. (A) RNA blot analysis of the expression of the five 2S genes in leaves of four sdg8 mutants grown for 14 d on MS agar. Wild-type (Col) leaves and siliques were used as negative and positive controls, respectively. The same amount of RNA was used for each blot. Elongation factor 1α was used as loading control. (B) Real-time quantitative RT-PCR (qRT-PCR) validation of the expression in sdg8-5 leaves of seed-related genes revealed in the DNA microarray analysis. RNAs from leaves of 14-day-old plants grown on MS agar were used for PCR. Only those validated by qRT-PCR are shown here. Wild-type (βCGpro:GUS) RNA levels are designed as 1-fold. The expression of Actin-8 was used as an internal control. The mean and standard error (SE) were determined from three biological replicates. Bars represent SEs. (C) qRT-PCR analysis of ABI3, FUS3, LEC1, and LEC2 genes in seedlings (aerial portion) of sdg8-2 mutants grown for 14 d on MS agar. Wild-type (Col) RNA levels are designed as 1-fold. The expression of Actin-8 was used as an internal control. The mean and SE were determined from three biological replicates, each of which was conducted in triplicate.