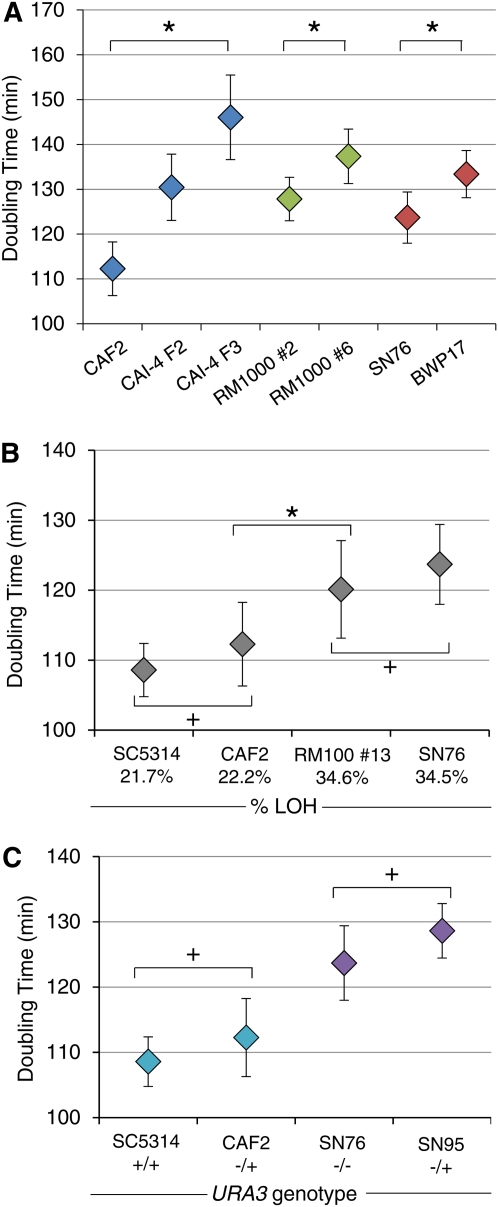
Figure 5 .
In vitro growth of lab strains with different degrees of ploidy or LOH or with different URA3 copy number. (A) Deviations from diploidy correlate with increased doubling times. Doubling times (min) were measured for strains with similar degrees of LOH and different degrees whole-genome ploidy. Blue: CAF2 (2N), CAI-4 F2 [2.16N (2N+Chr2)] and CAI-4 F3 [2.38 (2N + (Chr1 + Chr2))] Green: RM1000 #2 (2N) and RM1000 #6 (1.997N) have identical genome content but are homozygous or hemizygous, respectively, for the terminal ∼40 kb of Chr5R. Red: SN76 (2N) and BWP17 (1.997N), like the RM1000 strains, are homozygous and hemizygous for the tip of Chr5R; in addition, BWP17 acquired a small (30 kb) LOH on Chr4R. *Within each group, all strains are significantly different from each other (P < 0.05, Student t-test). (B) Increases in percentage of the genome that is homozygous (as indicated on X-axis) correlate with increased doubling times. SC5314 and CAF2 are not different from each other (+) but are significantly different from RM100 #13 and SN76 (*P < 0.05, Student t-test). (C) URA3 status does not affect in vitro doubling times in medium containing uridine. Pairs of strains (teal and purple) with similar ploidies and similar degrees of LOH that differ by one copy of URA3: SC5314 (URA3/URA3) vs. CAF2 (URA3/ura3∆), or SN76 (ura3∆/ura3∆) vs. SN95 (URA3/ura3)) were not significant different in doubling time relative to one another (+). LOH, loss ofheterozygosity.