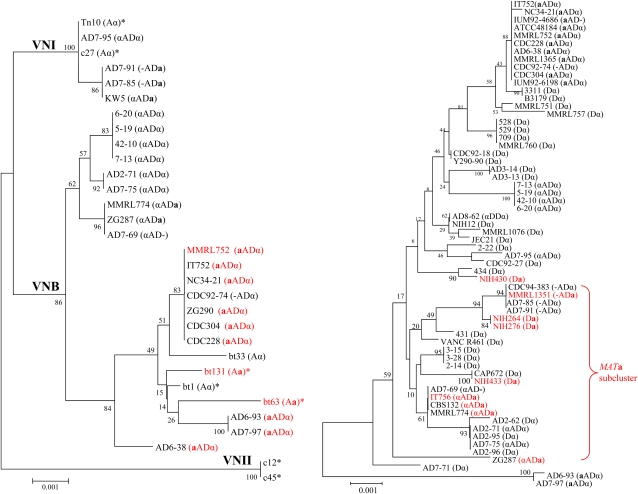
Figure 2 .
Phylogenetic organization of C. neoformans serotype A (left) and serotype D (right). This analysis was based on the concatenation of 5 MLST markers: IGS, URE1, GPD1, LAC1, and MPD1. The five loci were specifically amplified and sequenced from the serotype A and D genomes of the AD hybrid isolate by the use of serotype-specific primers. For comparison with haploid strains, 32 serotype D haploid strains, 1 serotype DD diploid strains, and 7 serotype A strains (*) of VNI, VNII, and VNB lineages from a previous study (Litvintseva et al. 2006) were included in the phylogenetic analysis. The concatenated DNA sequences were aligned by the use of ClustalW and MUSCLE. The phylogenetic organization was constructed with Mega 3.1 via the neighbor-joining method. The MATa strains are shown in red. The serotype and mating type of each isolate are indicated in brackets.