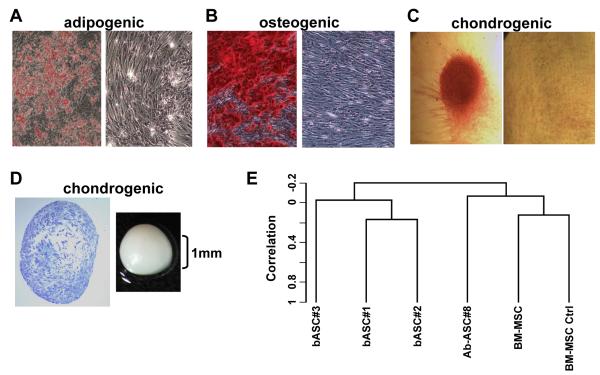
Figure 1. Breast-derived ASCs exhibit multipotent mesenchymal differentiation.
Breast ASCs were cultured in media to trigger adipogenic, osteogenic and chondrogenic lineages and then histochemically stained, as described previously [2], with: (A) Oil Red O to detect lipid droplets; (B) Alizarin red to detect extracellular calcium; and (C) Safranin O to detect glycosoaminoglycans where the micromass was removed for microscopic analysis. Undifferentiated breast ASCs were included as negative controls (right panels of A-C). (D) Cartilage micromass (gross image on right) and toludine staining of the sectioned micromass (left). (E) Dendrogram from unsupervised hierarchical clustering using centered correlation and average linkage. Four micrograms of total RNA from each sample was used to generate double-stranded cDNA using a 24-mer oligodeoxy-thymidylic acid primer with a T7 RNA polymerase promoter site added to the 3′ end [19]. Second strand cDNA synthesis, cleanup, and biotinylation were conducted according to a standard protocol. Ten μg of fragmented cRNA were then hybridized on the GeneChip® Human Genome U133A 2.0 (HG-U133A 2.0) array and analyzed as described previously [20]. Note: b: breast; Ab: abdominal; and the number appearing after the respective cell cultures reflect individual cell isolations from different donors.