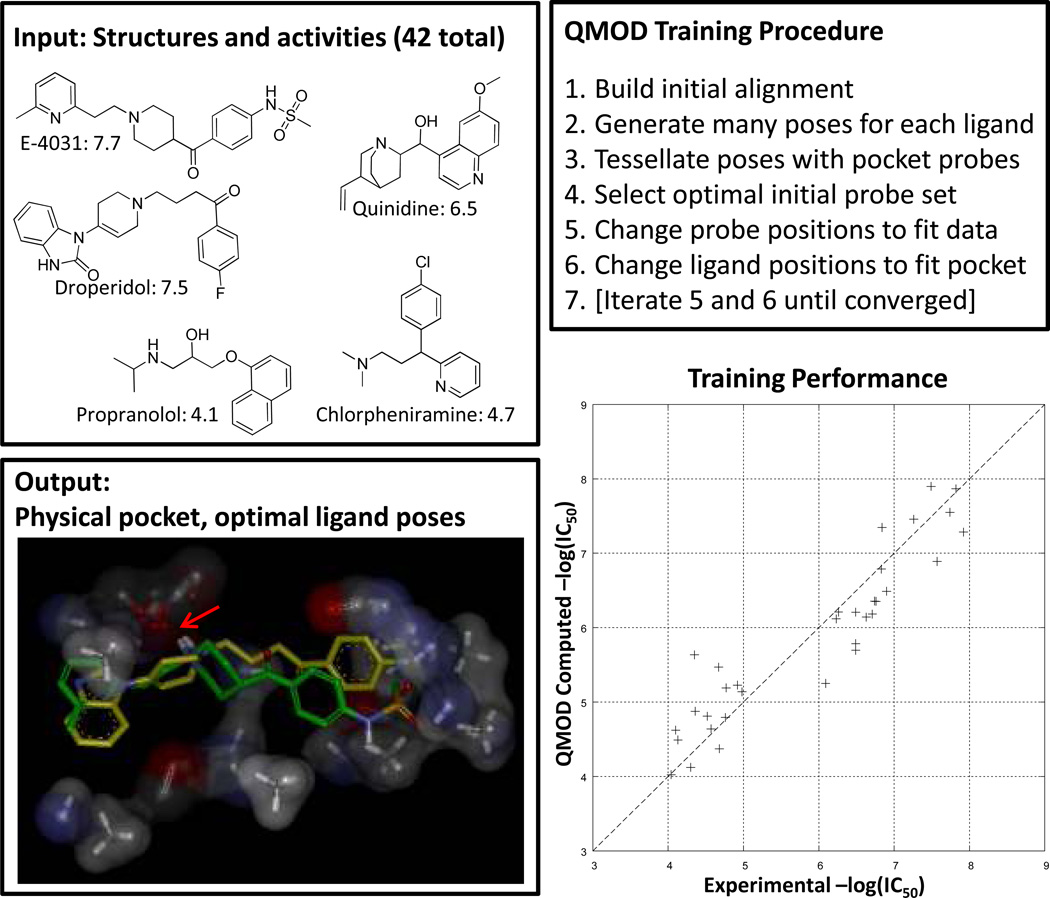
Figure 4.
Surflex-QMOD takes structures and activities as input and produces a physical model of a binding pocket. Training ligands in their optimal poses (according to the pocket’s preferences) produce scores close to those experimentally measured. The resulting model, by construction, yields a strong correlation with experimental activity (Kendall’s Tau = 0.80 (p << 0.001), r2 = 0.86). The final pocket along with the optimal poses of E-4031 (green) and droperidol (yellow) are shown at bottom left. The critical interaction with the ligands’ protonated amine is highlighted with a red arrow. The pocketmol has multiple carbonyl probes, which together approximate the effect of a negative charge of a particular magnitude, with some degree of mobility.