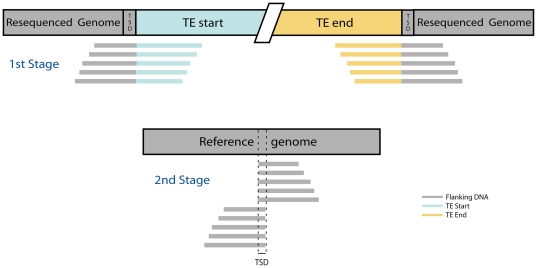
Figure 1. Overview of de novo TE insertion site mapping strategy.
We detected de novo TE insertions using a two-stage process that relies on the presence of TSDs. In the first stage (top), unaligned and unassembled WGS sequence reads from a resequenced genome that has an integrated TE insertion were queried against a library of canonical TE sequences. Reads that span the junction of the start or end of TE and genomic flanking sequences are retained. In the second stage (bottom), the unique genomic DNA components of junction reads identified previously were aligned against the reference genome. The region of overlap between sets of junction reads that span the start and end of the same TE was used to define the TSD and orientation of de novo TE insertions.