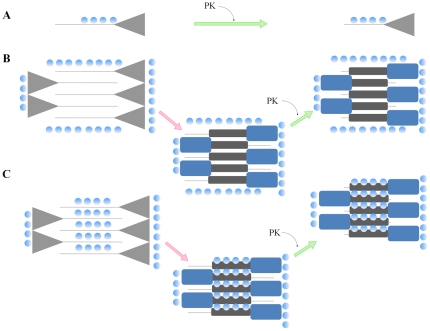
Figure 6. Potential mechanisms for recPrP aggregation induced by kosmotropic salts.
The soluble monomeric recPrP is represented as a two-domain protein, C-terminal globular domain (triangle) containing a large proportion of alpha-helical structure and the natively-unfolded N-terminal domain (solid line). Misfolded recPrP acquires intermolecular beta-sheet secondary structure in the natively unfolded region 90–145 (represented as a dark gray horizontal rectangle). The structural fate of the C-terminal domain is unclear, but most likely involves a conformational rearrangement (represented as a dark blue rectangle). The kosmotropic anions are represented by the blue circles. The dotted arrows indicate a misfolding process, while the solid arrows represent the protease digestion reaction (PK). Three putative models to explain the effect of the salt on inducing the formation of recPrPres are proposed. A: Binding of kosmotropic anions may occlude protease cleavage sites within the N-terminal domain of PrP. However, this model per se does not account for the partial resistance to proteolytic degradation of the whole C-terminal domain as well as for the structural changes induced by salt. B : A salting-out-like mechanism locally increases PrP concentration in a native-like conformation, followed by either an induction or acceleration of misfolding to form intermolecular beta-sheets giving rise to PrPSc-like aggregates. C : A combined effect of high concentrations of kosmotropic anions that partially salt-out recPrP in a close-to-native fold from the bulk solution, along with specific anion binding to PrP that further stabilizes the N-terminal domain induce a protease-resistant recPrP conformation with PrPSc features.