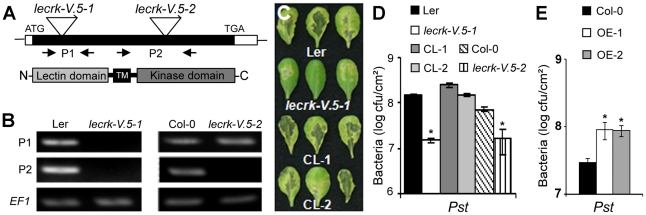
Figure 1. LecRK-V.5 negatively regulates disease resistance to bacteria.
(A) Insertional mutation sites in two lecrk-V.5 mutant lines. Ds transposon (lecrk-V.5-1) and T-DNA (lecrk-V.5-2) insertion sites are shown. Filled box represents exon and arrows denote the different positions of the primers used in the RT-PCR experiments in (B). The relative position of lectin, transmembrane (TM) and kinase domains in LecRK-V.5 predicted protein structure is indicated. (B) RT-PCR analysis of LecRK-V.5 transcripts in WT and lecrk-V.5 mutants. EF-1 was used as a control. (C) Disease symptoms assessed 3 days after dip-inoculation with 1×107 cfu.ml−1 Pst DC3000 in WT (Ler), lecrk-V.5-1 and two complemented lines (CL-1 and CL-2). (D) Bacterial growth at 3 days post-inoculation (1×107 cfu.ml−1 Pst DC3000 (Pst)) in WT (Col-0 and Ler), lecrk-V.5 mutants and two complemented lines (CL-1 and CL-2). (E) Bacterial titers evaluated 3 days after dip-inoculation with 1×107 cfu.ml−1 Pst DC3000 (Pst) in lines overexpressing LecRK-V.5 (OE-1 and OE-2). For (D) and (E), data represent average ± SD. Statistical differences between WT controls and mutants or transgenics are detected with a t test (P<0.01, n = 6). All experiments were repeated at least three times with similar results.