Figure 3.
Predicted Structural Effects of MFDM-Causing Missense Mutations
We modeled residues 114–957 of EFTUD2 on the crystal structure of S. cerevisiae ribosomal elongation factor 2 (eEF2)14 by using ModWeb, and we visualized them in PyMol.
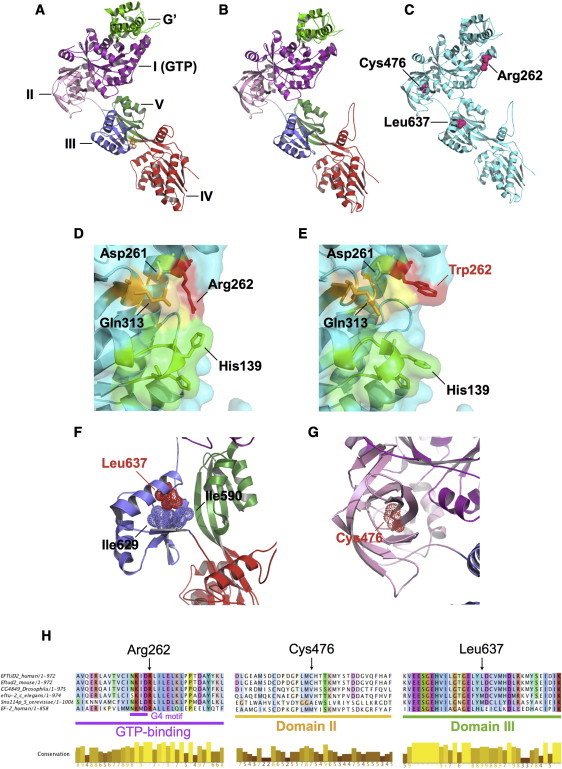
(A) Crystal structure of eEF2 (PDB: 1N0U) complexed with the translation inhibitor sordarin (yellow). Structural domains are highlighted.
(B) U5-116kD model based on eEF2. The modeled portion of U5-116kD has high identity (37%) with eEF2; ModWeb scores this model as “reliable” (GA341 score is 1.00).
(C) Locations of amino acid substitutions p.Arg262Trp, p.Cys476Arg, and p.Leu637Arg.
(D) Arg262 (red) is predicted to occupy surface of GTP binding site. Also highlighted are additional surface-facing residues of G1 (green), G4 (yellow), and G5 (orange) motifs.
(E) Arg262Trp alteration in individual 2 is predicted to alter GTP-binding surface topology.
(F) Leu637 sidechain (helix II of domain III) is predicted to face inward and to form close hydrophobic contacts with Ile590 and Ile629. Leu637Arg (individual 11) is predicted to introduce a positive charge at this location.
(G) Cys476 is situated at the center of domain II's β barrel structure. p.Cys476Arg (individual 12) is predicted to introduce a positive charge at this position.
(H) Multispecies alignments of U5-116kDa illustrate sequence conservation in the vicinity of missense mutations identified in individuals with MFDM.
