Figure 2.
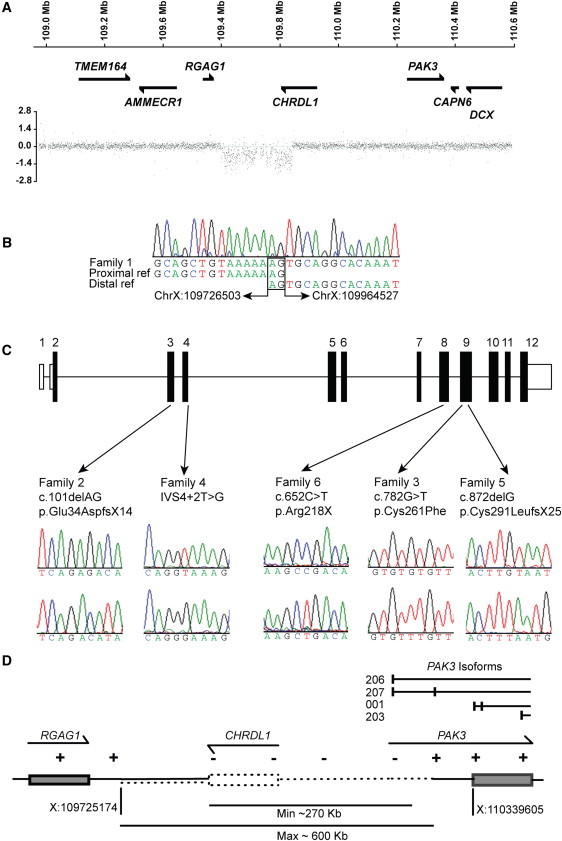
CHRDL1 Mutations Cause X-Linked Megalocornea
(A) X chromosome dense array comparative genomic hybridization for family 1 revealed a deletion within the disease interval of approximately 240 kb that encompassed the 3′ end of CHRDL1. Genes are schematically represented against the position on the X chromosome.
(B) The deletion breakpoint sequence is shown with chromosome X:109726503 joined to sequence chromosome X:109964527 sequence (238,024 bp deleted; hg19 genome build). Microhomology at the deletion breakpoints is highlighted (2 bp; AG).
(C) Schematic of CHRDL1 with patient mutations identified in families 2–6. Nonsense, frameshift, splicing, and missense mutations were identified in exons 3, 4, 8, and 9. Patient sequence electropherograms are shown above control sequence electropherograms.
(D) Schematic of segmental deletion identified in family 7. The deletion encompasses the entire CHRDL1 gene and extends distally to PAK3. The first untranslated exon of PAK3 isoforms 206 and 207 (Ensembl nomenclature hg19 genome build) is also deleted.
