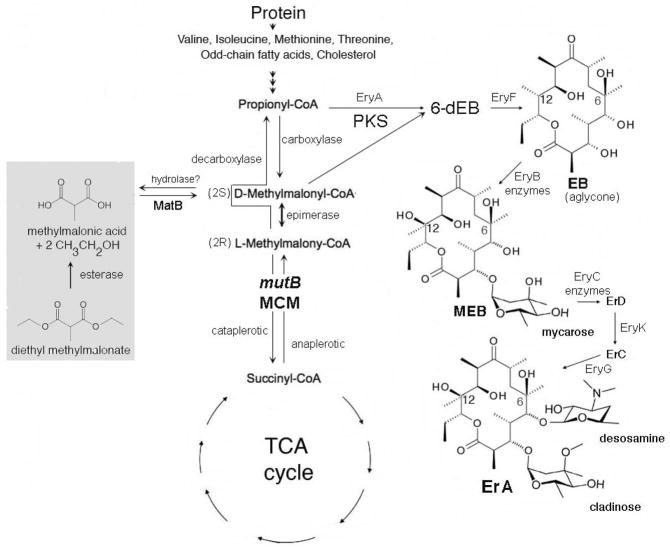
Fig. 1.
Metabolic map of the propionate pathway and erythromycin biosynthetic pathway. Hypothetical pathway (gray box) shows diethyl methylmalonate assimilation into erythromycin (Lombo et al. 2001). A hydrolase, converting methylmalonyl-CoA to methylmalonic acid is also postulated. Abbreviations: MCM, methylmalonyl-CoA mutase; EB, erythronolide B; 6-dEB, 6-deoxyerythronolide B; MEB, 3-O-α-mycarosylerythronolide B; ErA, C, and D, erythromycin A, C and D. EryA, polyketide synthase; EryF, C-6 hydroxylase; EryB, dTDP-mycarose biosynthetic and transferase enzymes; EryC, dTDP-desosamine biosynthetic and transferase enzymes; EryK, C-12 hydroxylase; EryG, S-adenosylmethionine methylase acting on the attached mycarose to convert it to cladinose.