Abstract
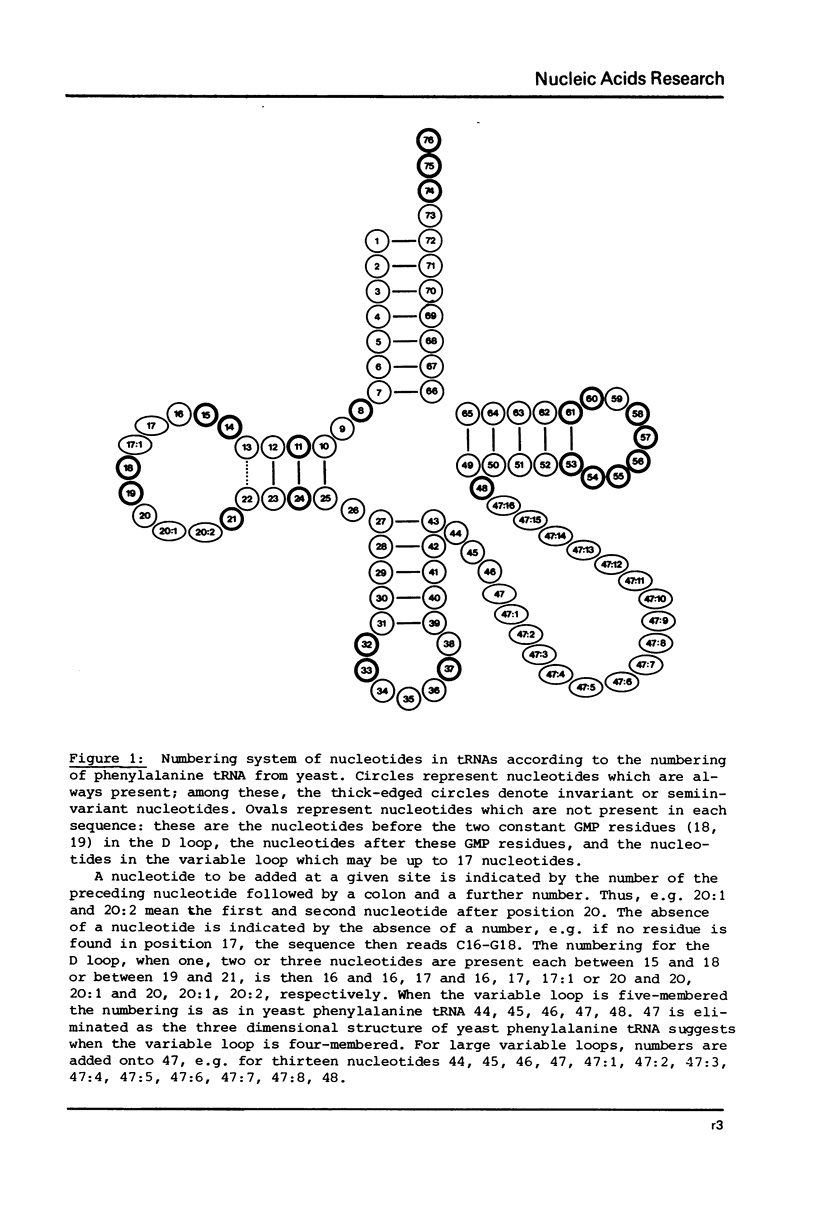
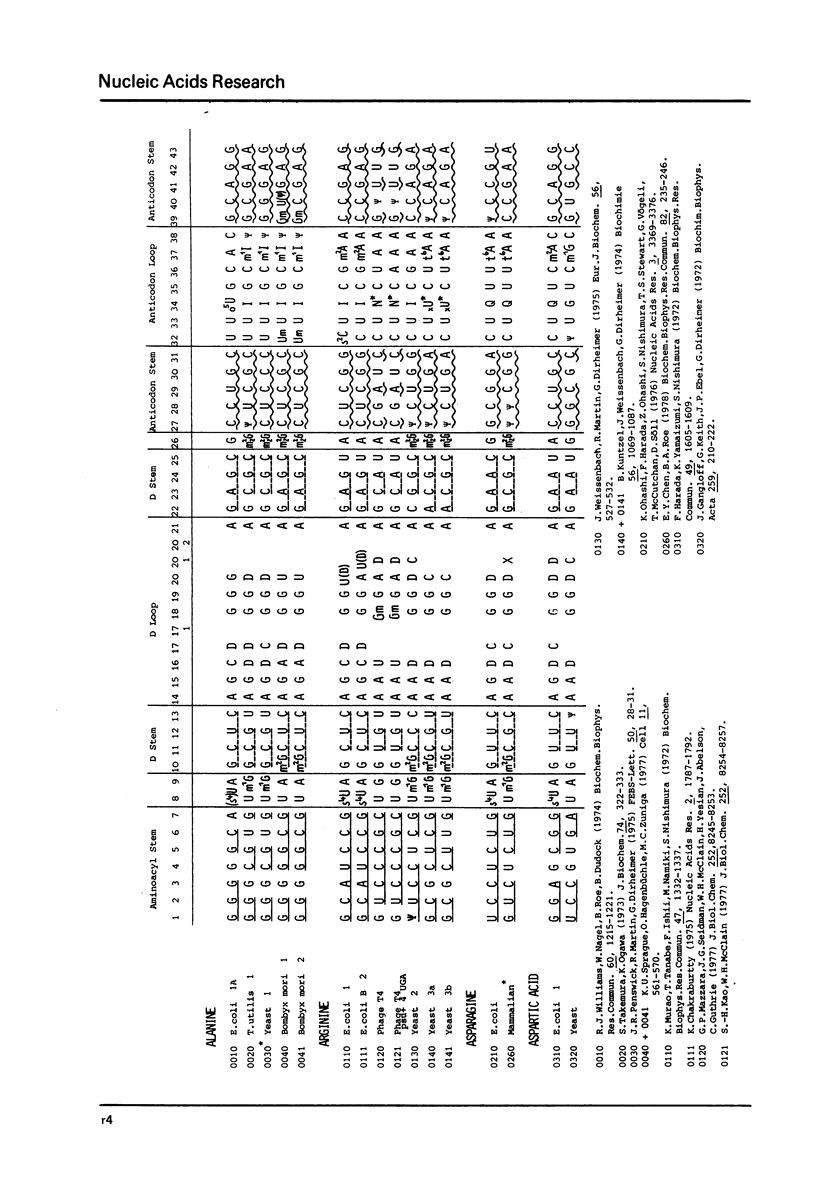
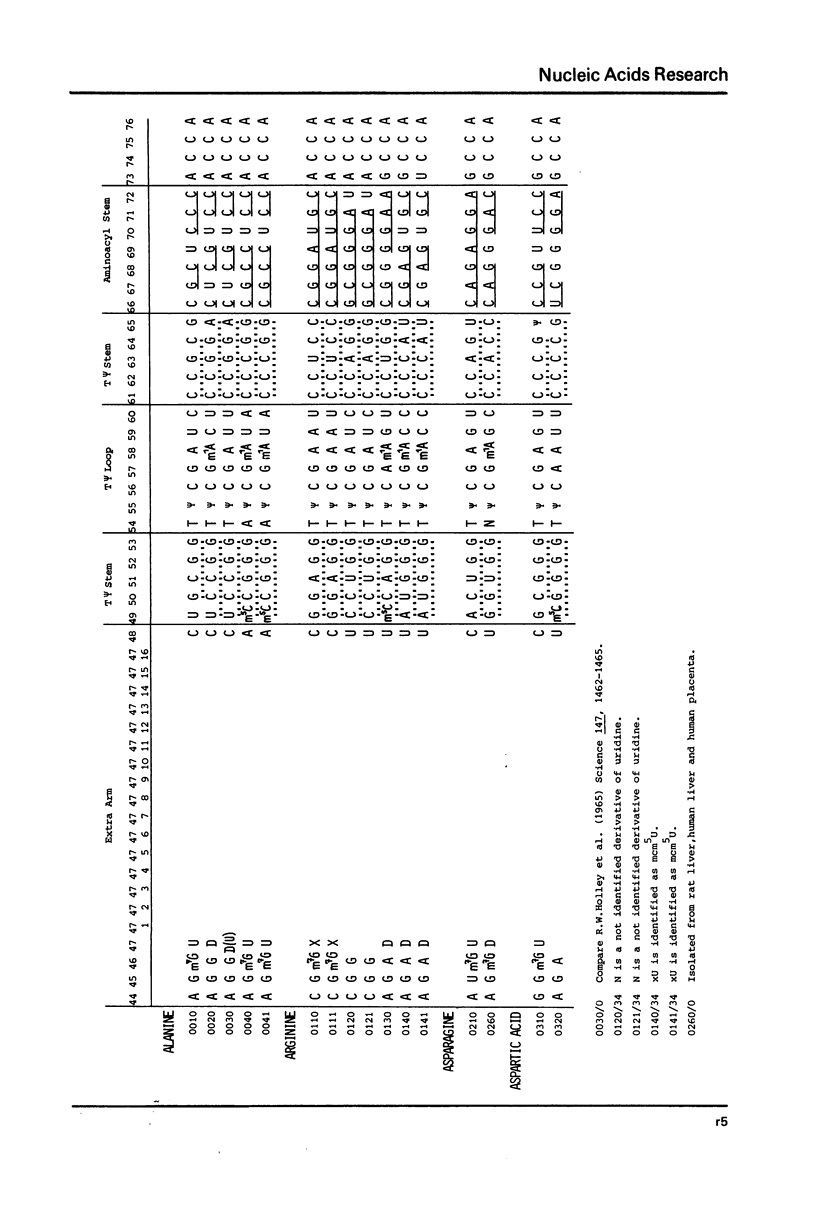
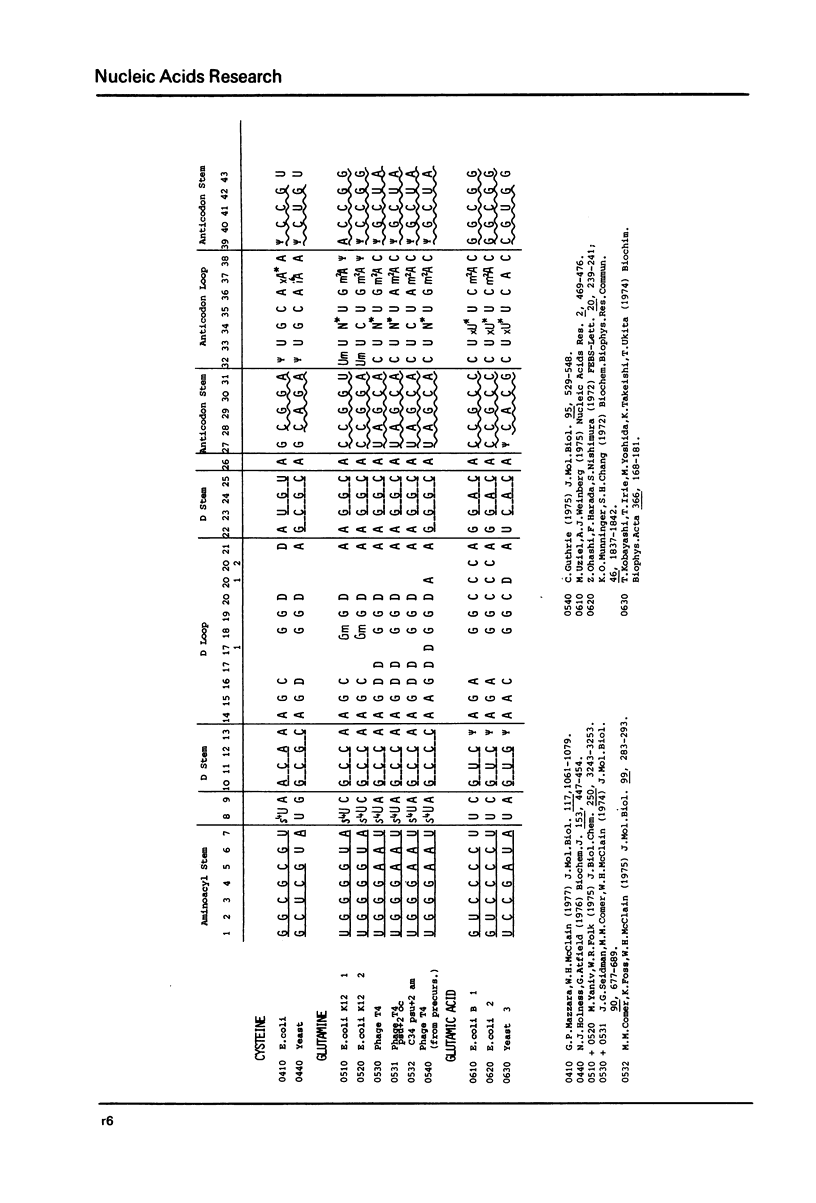
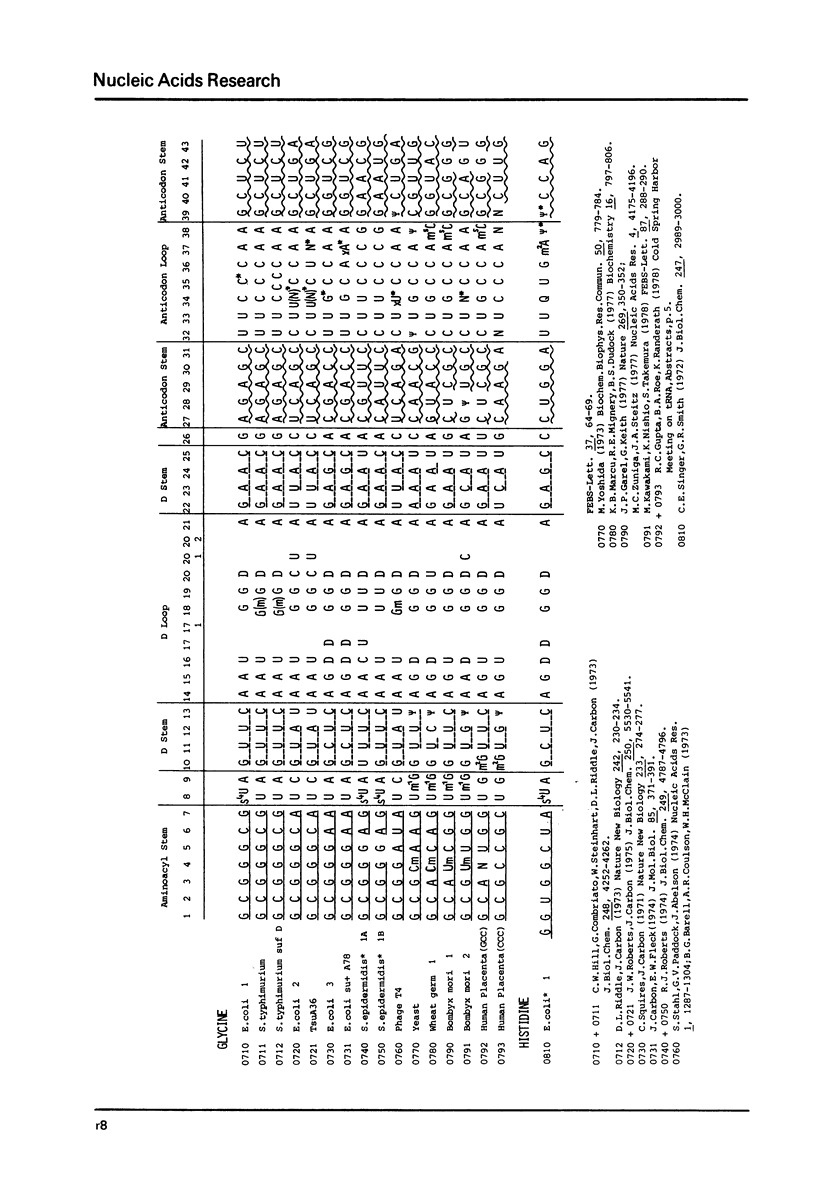
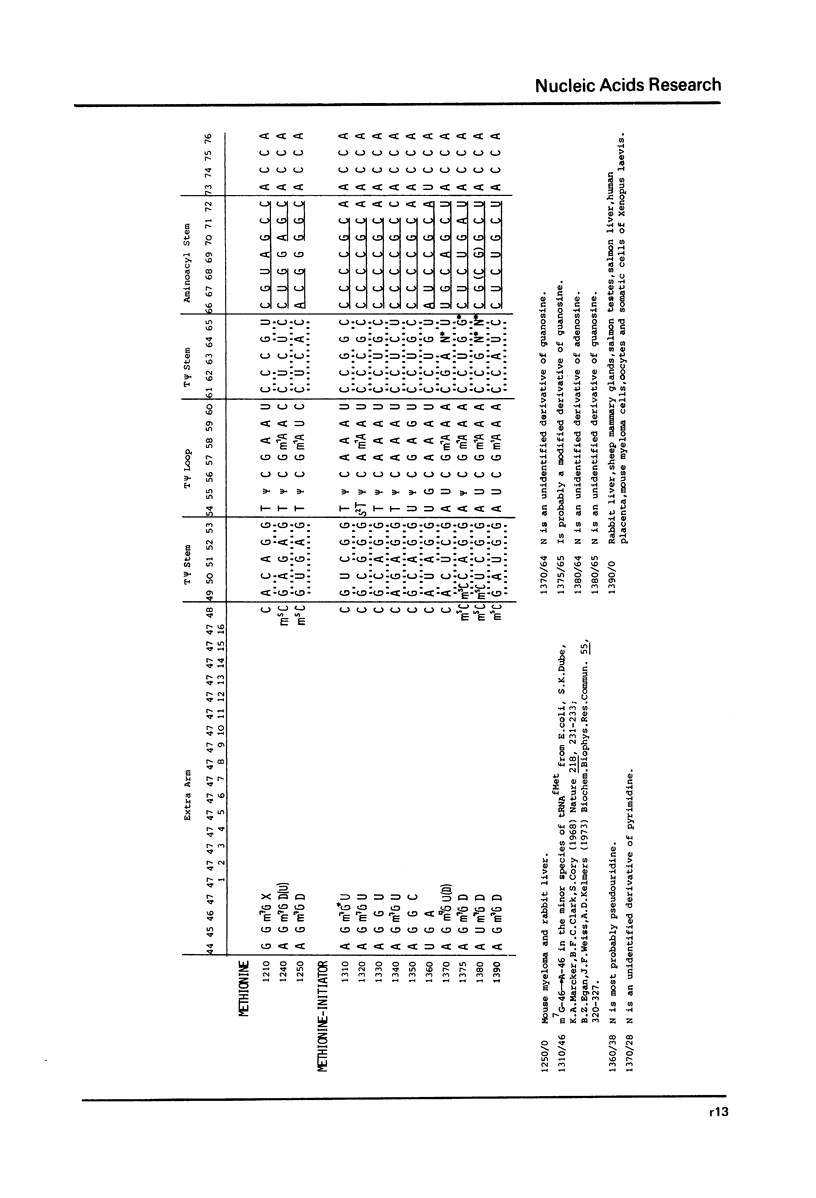
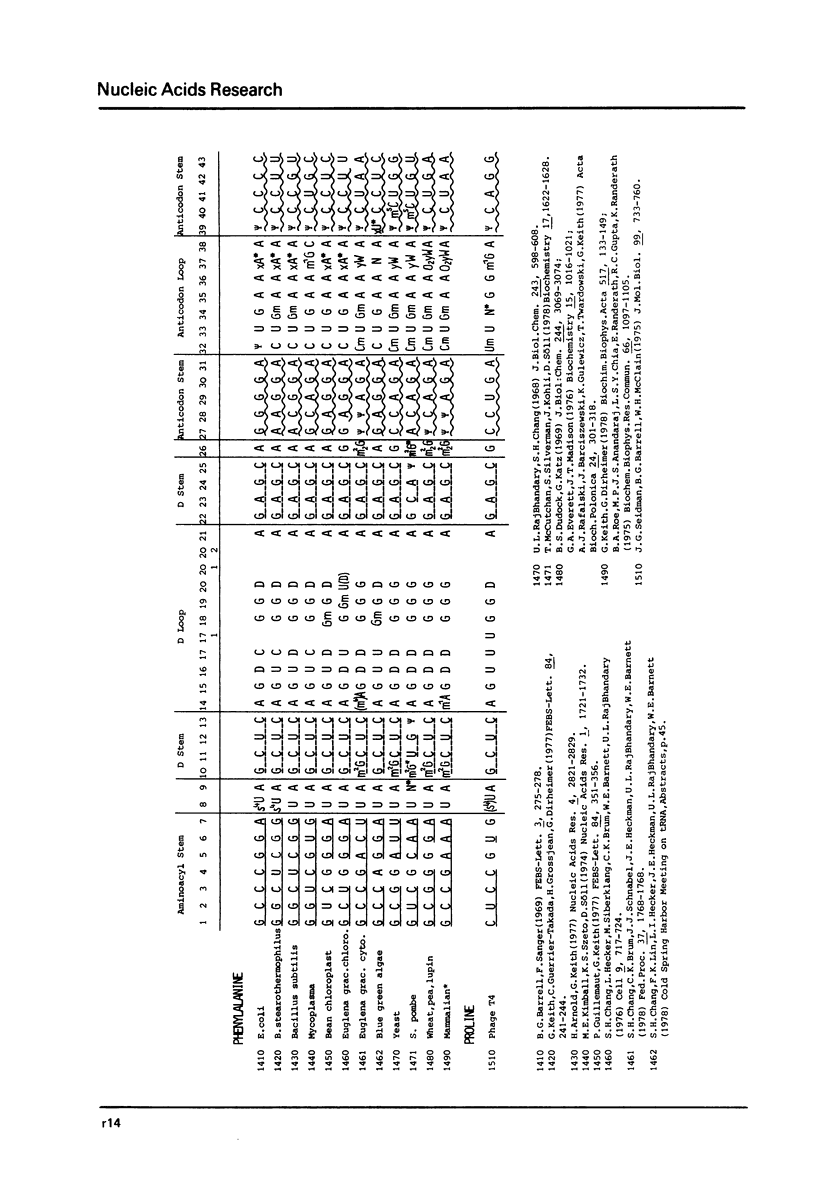
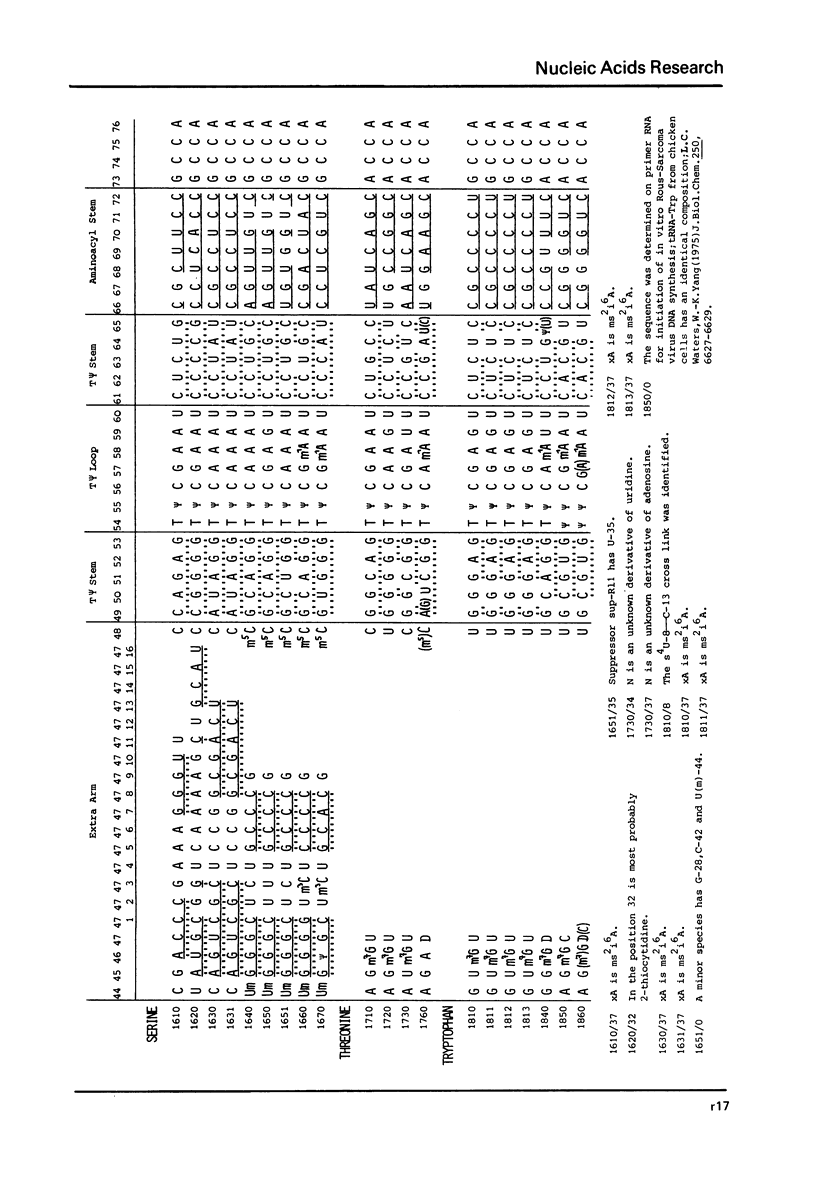
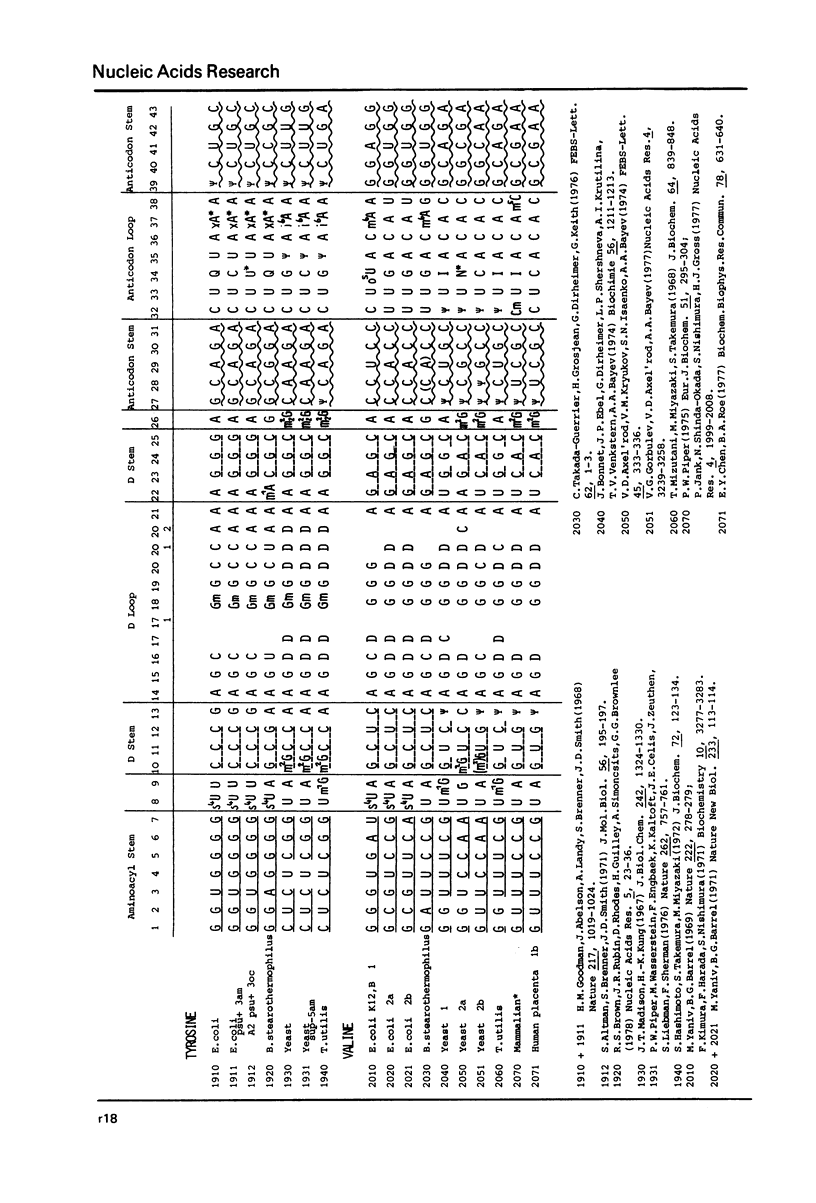
This compilation presents in a small space the tRNA sequences so far published in order to enable rapid orientation and comparison. The numbering of tRNAPhe from yeast is used as has been done earlier (1) but following the rules proposed by the participants of the Cold Spring Harbor Meeting on tRNA 1978 (2) (Fig. 1). This numbering allows comparisons with the three dimensional structure of tRNAPhe, the only structure known from X-ray analysis. The secondary structure of tRNAs is indicated by specific underlining. In the primary structure a nucleoside followed by a nucleoside in brackets or a modification in brackets denotes that both types of nucleosides can occupy this position. Part of a sequence in brackets designates a piece of sequence not unambiguously analyzed. Rare nucleosides are named according to the IUPAC-IUB rules (for some more complicated rare nucleosides and their identification see Table 1); those with lengthy names are given with the prefix x and specified in the footnotes. Footnotes are numbered according to the coordinates of the corresponding nucleoside and are indicated in the sequence by an asterisk. The references are restricted to the citation of the latest publication in those cases where several papers deal with one sequence. For additional information the reader is referred either to the original literature or to other tRNA sequence compilations (3--7). Mutant tRNAs are dealt with in a separate compilation prepared by J. Celis (see below). The compilers would welcome any information by the readers regarding missing material or erroneous presentation. On the basis of this numbering system computer printed compilations of tRNA sequences in a linear form and in cloverleaf form are in preparation.
Full text
PDF









Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Dirheimer G., Ebel J. P., Bonnet J., Gangloff J., Keith G., Krebs B., Kuntzel B., Roy A., Weissenbach J., Werner C. Structure primaire des tRN. Biochimie. 1972;54(2):127–144. doi: 10.1016/s0300-9084(72)80097-x. [DOI] [PubMed] [Google Scholar]
- Feinberg A. M., Nakanishi K., Barciszewski J., Rafalski A. J., Augustyniak H., Wiewiórowski M. Isolation and characterization of peroxy-Y base from phenylalanine transfer ribonucleic acid of the plant, Lupinus luteus. J Am Chem Soc. 1974 Dec 11;96(25):7797–7780. doi: 10.1021/ja00832a029. [DOI] [PubMed] [Google Scholar]
- Feldman M. Y. Minor components in transfer RNA: the location-function relationships. Prog Biophys Mol Biol. 1977;32(1):83–102. [PubMed] [Google Scholar]
- Friedman S., Li H. J., Nakanishi K., Van Lear G. 3-(3-amino-3-carboxy-n-propyl)uridine. The structure of the nucleoside in Escherichia coli transfer ribonucleic acid that reacts with phenoxyacetoxysuccinimide. Biochemistry. 1974 Jul 2;13(14):2932–2937. doi: 10.1021/bi00711a024. [DOI] [PubMed] [Google Scholar]
- Goddard J. P. The structures and functions of transfer RNA. Prog Biophys Mol Biol. 1977;32(3):233–308. [PubMed] [Google Scholar]
- Harada F., Gross H. J., Kimura F., Chang S. H., Nishimura S., RajBhandary U. L. 2-Methylthio N6-(delta 2-isopentenyl) adenosine: a component of E. coli tyrosine transfer RNA. Biochem Biophys Res Commun. 1968 Oct 24;33(2):299–306. doi: 10.1016/0006-291x(68)90784-5. [DOI] [PubMed] [Google Scholar]
- Kasai H., Oashi Z., Harada F., Nishimura S., Oppenheimer N. J., Crain P. F., Liehr J. G., von Minden D. L., McCloskey J. A. Structure of the modified nucleoside Q isolated from Escherichia coli transfer ribonucleic acid. 7-(4,5-cis-Dihydroxy-1-cyclopenten-3-ylaminomethyl)-7-deazaguanosine. Biochemistry. 1975 Sep 23;14(19):4198–4208. doi: 10.1021/bi00690a008. [DOI] [PubMed] [Google Scholar]
- Kuntzel B., Weissenbach J., Wolff R. E., Tumaitis-Kennedy T. D., Lane B. G., Dirheimer G. Presence of the methylester of 5-carboxymethyl uridine in the wobble position of the anticodon of tRNAIII Arg from brewer's yeast. Biochimie. 1975;57(1):61–70. doi: 10.1016/s0300-9084(75)80110-6. [DOI] [PubMed] [Google Scholar]
- Nakanishi K., Furutachi N., Funamizu M., Grunberger D., Weinstein I. B. Structure of the fluorescent Y base from yeast phenylalanine transfer ribonucleic acid. J Am Chem Soc. 1970 Dec 30;92(26):7617–7619. doi: 10.1021/ja00729a035. [DOI] [PubMed] [Google Scholar]
- Nishimura S., Taya Y., Kuchino Y., Oashi Z. Enzymatic synthesis of 3-(3-amino-3-carboxypropyl)uridine in Escherichia coli phenylalanine transfer RNA: transfer of the 3-amino-acid-3-carboxypropyl group from S-adenosylmethionine. Biochem Biophys Res Commun. 1974 Apr 8;57(3):702–708. doi: 10.1016/0006-291x(74)90603-2. [DOI] [PubMed] [Google Scholar]
- Ohashi Z., Maeda M., McCloskey J. A., Nishimura S. 3-(3-Amino-3-carboxypropyl)uridine: a novel modified nucleoside isolated from Escherichia coli phenylalanine transfer ribonucleic acid. Biochemistry. 1974 Jun 4;13(12):2620–2625. doi: 10.1021/bi00709a023. [DOI] [PubMed] [Google Scholar]
- Sodd M. A., Doctor B. P. Nucleotide sequences of transfer ribonucleic acids. Methods Enzymol. 1974;29:741–756. doi: 10.1016/0076-6879(74)29066-9. [DOI] [PubMed] [Google Scholar]
- Sprinzl M., Grüter F., Gauss D. H. Collection of published tRNA sequences. Nucleic Acids Res. 1978 May;5(5):r15–r27. [PMC free article] [PubMed] [Google Scholar]
- Yamada Y., Nishimura S., Ishikura H. The presence of 2-methylthio-N 6 -( 2 -isopentenyl)adenosine in leucine, tryptophan and cysteine tRNA's from Escherichia coli. Biochim Biophys Acta. 1971 Sep 30;247(1):170–174. doi: 10.1016/0005-2787(71)90821-5. [DOI] [PubMed] [Google Scholar]


