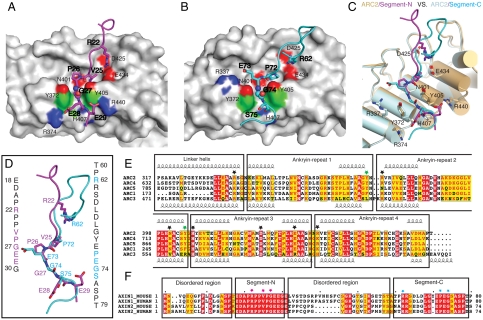
Fig. 2.
Structural details of the mouse TNKS1(308-655)/Axin1(1-80) interactions. (A) TNKS1-Axin1 segment-N interface. One copy of ARC2 of TNKS is shown in gray solid surface. Segment-N of Axin1 is shown in stick model in magenta. Residues involved in the interface are labeled. Regular font is used for labeling TNKS1 residues and bold font for Axin1 residues. The critical glycine is shown in sphere mode. (B) TNKS1-Axin1 segment-C interface. Another copy of ARC2 of TNKS is shown in gray solid surface as in A. Segment-C of Axin1 is shown in stick model in cyan. Residues are labeled in the same way as in A. (C) Superpositon of two TNKS1 ARC2-Axin1 interfaces. Two ARC2s are shown in cylindrical cartoon in light pink (interacting with Segment-N) and light gray (interacting with Segment-C). Residues of TNKS in the interface are labeled. The distance between the two parallel tyrosine side chains is approximately 7.3 Å. (D) Superposition of the two Axin1 segments. Color coding is the same as in (A) and (B). Sequences of two segments are shown in vertical text with critical residues colored in magenta and cyan, respectively. (E) Sequence alignment of five ARCs in mouse TNKS1. Individual ankyrin repeats as well as the linker helices between ARCs are boxed and labeled. Critical residues for the interaction are highlighted with black stars. The two gate-forming tyrosines (Tyr-372 and Tyr-405) are highlighted with green stars. Tyr-405 side chain also forms a hydrogen bond with the Axin mainchain. (F) Sequence alignment of the TNKS-binding domains of human and mouse Axin1/Axin2. Segment-N, Segment-C as well as the disordered region in the structure are boxed and labeled. Key residues from Segment-N and Segment-C are highlighted with red and cyan stars, respectively.