Abstract
For many model organisms traditionally in use for cardiac electrophysiological studies, characterization of ion channel genes is lacking. We focused here on two genes encoding the inward rectifier current, KCNJ2 and KCNJ12, in the dog heart. A combination of RT-PCR, 5′-RACE, and 3′-RACE demonstrated the status of KCNJ2 as a two exon gene. The complete open reading frame (ORF) was located on the second exon. One transcription initiation site was mapped. Four differential transcription termination sites were found downstream of two consensus polyadenylation signals. The canine KCNJ12 gene was found to consist of three exons, with its ORF located on the third exon. One transcription initiation and one termination site were found. No alternative splicing was observed in right ventricle or brain cortex. The gene structure of canine KCNJ2 and KCNJ12 was conserved amongst other vertebrates, while current GenBank gene annotation was determined as incomplete. In silico translation of KCN12 revealed a non-conserved glycine rich stretch located near the carboxy-terminus of the KIR2.2 protein. However, no differences were observed when comparing dog with human KIR2.2 protein upon ectopic expression in COS-7 or HEK293 cells with respect to subcellular localization or electrophysiological properties.
Keywords: inward rectifier current, Canis familiaris, KCNJ2, KCNJ12, KIR2.1, KIR2.2, genes
Introduction
In the mammalian heart, the resting membrane potential of cardiomyocytes is set and stabilized by the inward rectifier potassium current (IK1; Dhamoon and Jalife, 2005). In addition, IK1 contributes to outward potassium current during the last phase of action potential repolarization. The main molecular determinants of cardiac IK1 are the KIR2.1 and KIR2.2 proteins expressed from the KCNJ2 and KCNJ12 genes respectively (De Boer et al., 2010a). Defective inward rectifier current may lead, amongst other features, to lethal cardiac arrhythmias in mice and man such as ventricular arrhythmias and atrial fibrillation (Anumonwo and Lopatin, 2010). ECG recording from neonatal mice homozygous for a KCNJ2 null mutation showed lengthening of RR, PR, and QT intervals and QRS broadening. Furthermore, isolated neonatal cardiomyocytes displayed action potential lengthening and ectopic activity (Zaritsky et al., 2001). In contrast, however, null mutation of KCNJ12 generated no cardiac abnormalities (Zaritsky et al., 2001). KCNJ2 loss of function mutation associated with Andersen–Tawil syndrome 1 regularly, but not in each case, displayed long repolarization times (LQT) and biventricular tachycardias. On the other hand, KCNJ2 gain of function mutations have been associated with short QT (Priori et al., 2005), and atrial fibrillation (Xia et al., 2005). Atrial fibrillation was also observed in a mouse model overexpressing KCNJ2 (Li et al., 2004). Finally, KCNJ2 mutations have been associated with Catecholaminergic Polymorphic Ventricular Tachycardia (Vega et al., 2009). Affected KIR2.1 and KIR2.2 functioning may be the result of amino-acid substitutions (Tristani-Firouzi and Etheridge, 2010), direct channel block (Rodríguez-Menchaca et al., 2008; De Boer et al., 2010b), or changes in expression regulation (Yang et al., 2007).
The dog (Canis lupus familiaris, Cf) with chronic complete atrial–ventricular block (cAVB) is a well established model for drug-induced arrhythmia (Thomsen et al., 2006a). The model acquires its sensitivity and specificity from bradycardia associated volume overload and subsequent cardiac remodeling (Thomsen et al., 2007). The latter process translates into modified contractile, structural, and electrical function (Oros et al., 2008). As a result of electrical remodeling, the so-called repolarization reserve (Roden, 1998; Michael et al., 2009) is diminished as evidenced by increased action potential duration and moreover by increased beat-to-beat variation of repolarization (Thomsen et al., 2006b).
For some species, the molecular basis underlying cardiac electrophysiology, e.g., ion channel, gap junction, and transporter genes and proteins, is well described. However, a functional integration of the individual components to explain a number of electrophysiological phenomena lags behind, despite many decades of research (Coronel, 2010). When considering the dog, the amount of electrophysiological studies by far outnumbers those on the molecular make-up of the underlying ion currents. Nevertheless, the canine genes coding for the electrophysiological building blocks are becoming deciphered rapidly; genome project information can be found at http://www.broadinstitute.org/mammals/dog. The publication of the dog genome has provided an important additional tool that stimulated molecular research in this model species (Lindblad-Toh et al., 2005). However, as algorithm driven gene annotation is improving gradually (Flicek, 2007), its results are still often doubtful indicating that experimental confirmation of the genomic structure of the genes of interest is required. To enable our future studies on KCNJ2 and KCNJ12 expression regulation in the dog, it is crucial to characterize the molecular structure of the respective genes. Here we have characterized the canine KCNJ2 and KCNJ12 gene structures by combining genome project data, RT-PCR, and 5′- and 3′-RACE protocols. We demonstrated that the canine KCNJ2 gene consists of two exons harboring its open reading frame (ORF) on exon 2. In contrast, we provided experimental evidence that KCNJ12 is a three exon gene. Its ORF is located on exon 3. No alternative splicing was detected in either heart or brain tissue. We demonstrated that the gene structure is conserved. Canine KIR2.2 harbors a peculiar stretch of glycine residues in its carboxy-terminus that is not present in KIR2.2 proteins from other species, which did not affect however its expression in cell systems neither its electrophysiological properties.
Materials and Methods
RT-PCR, 3′- and 5′-RACE
The investigation conformed to the Guide for the Care and Use of Laboratory Animals published by the US National Institutes of Health (NIH Publication No. 85-23, revised 1996) and was approved by the institutional committee for animal experiments. For all sequencing, samples from at least two purpose bred mongrel dogs were used, that harbored no known genetic defects.
Total RNA was isolated from canine ventricular cells or brain cortex tissue using Trizol (Invitrogen, Breda, The Netherlands) according to the manufacturers recommendations. mRNA was reverse transcribed using oligo dT and Superscript 2 (Invitrogen). Primer sequences for PCR are given in Table 1. Products were analyzed on ethidium-bromide stained 0.7–1.5% agarose gels. PCR products were isolated from gel using QIAquick Gel Extraction kit (Qiagen, Venlo, The Netherlands), cloned in pGEM-T-easy (Promega, Leiden, The Netherlands) and subsequently sequenced.
Table 1.
Sequences of primers used in this study, sequences are given in 5′–3′ orientation.
| KCNJ2 PRIMERS | |
| KCNJ2#1se | AGCTGGGTCTTGGGGATTCTGG |
| KCNJ2#2se | TTGCAGAGCGCACTGGAGCC |
| KCNJ2#3as | CATCCACCGCCAGCGAATGT |
| KCNJ2#4as | ACTGGACGTTGCAGTGGCCG |
| KCNJ2#5se | TCCGAGGAGACTCGCCGTGAAT |
| KCNJ2#6se | AGCAGAGTGAGCTCTCCTTCGC |
| KCNJ12 PRIMERS | |
| KCNJ12#1se | GCTGCTGCTGACACAGCCTTG |
| KCNJ12#2se | CGGGGTGTCCAGAGACCTGGGTTC |
| KCNJ12#3se | GGACCTGGAGACCGACGACTTCG |
| KCNJ12#4se | GAGCCGCGATGAGGAAGACGAGG |
| KCNJ12#5se | GGGTTTGAGCAGAATGGGCCTGG |
| KCNJ12#6as | GATCACCCAGAAGATGATGCC |
| KCNJ12#7as | GTTGCCGAAGCCGTTGGCACCCGAC |
| KCNJ12#8as | CCCGCCTGTGTCAGCAGCAGC |
| KCNJ12#9as | CGATGGTGGTCTGCGTCTCAATGG |
| KCNJ12#10se | GTGACCGATTCCTCTCCAGCT |
| KCNJ12#11se | GCAGGAAATGGAAGCTGCTGC |
| KCNJ12#12as | ACGCTACTGAGTCTGCAACCG |
| KCNJ13#13as | CCAGGCCCATTCTGCTCAAAC |
| SUPPLIED GENERACER™ PRIMERS USED | |
| GeneRacer™ 5′ RNA primer | CGACUGGAGCACGAGGACACUGACAUGGACUGAAGGAGUAGAAA |
| GeneRacer™ Oligo dT primer | GCTGTCAACGATACGCTACGTAACGGCATGACAGTG(T)24 |
| GeneRacer™ 5′ primer | CGACTGGAGCACGAGGACACTGA |
| GeneRacer™ 5′ nested primer | GGACACTGACATGGACTGAAGGAGTA |
| GeneRacer™ 3′ primer | GCTGTCAACGATACGCTACGTAACG |
| GeneRacer™ 3′ nested primer | CGCTACGTAACGGCATGACAGTG |
3′- and 5′-rapid amplification of cDNA ends (RACE) was performed on total RNA using GeneRacer™ kit (Invitrogen) according to the manufacturers recommendations. In short, total RNA was dephosphorylated and decapped followed by ligation of GeneRacer™ 5′ RNA primer. Subsequently, reverse transcription was performed using GeneRacer™ Oligo dT primer and Superscript 3. Finally, 5′ and 3′ RACE PCR was performed using KCNJ2 and 12 primers as indicated in the results and discussion section, in combination with primers supplied and recommended by the manufacturer. Sequences of primers used for RACE are depicted in Table 1. All PCR products were sequenced by the Dye-terminator sequencing method (BigDye®, Applied Biosystems, Nieuwerkerk a.d. IJssel, The Netherlands) on a 3730 Genetic Analyzer (Applied Biosystems).
Cloning and expression analysis of CfKCNJ12
Complete CfKCNJ12 coding sequence was amplified as above using primers KCNJ12#10se and KCNJ12#12as as first and KCNJ12#11se and KCNJ12#13as as nested primers, and cloned in pGEM-T-Easy followed by subcloning in pcDNA3.1 using EcoRI. COS-7 cells were transfected with pcDNA–CfKCNJ12 or pcDNA–HsKCNJ12 (human KCNJ12) construct using Lipofectamine 2000 (Invitrogen) according to the manufacturers recommendations. Twenty-four hours post-transfection, cells were harvested in lysis buffer [20 mM HEPES, pH 7.6, 125 mM NaCl, 10% (v/v) glycerol, 1 mM EDTA, 1 mM EGTA, 1 mM Dithiothreitol, 1% (v/v) Triton X-100]. Subsequently, 20 μg protein lysate was separated by 10% SDS-PAGE and blotted onto nitrocellulose membrane. Blots were blocked with 5% (v/v) fresh chicken egg yolk in TBST [20 mM Tris–Cl, pH 8.0, 150 mM NaCl, 0.05% (v/v) Tween-20] for 1 h at room temperature. KIR2.2 protein was detected using KIR2.1/2 antibody (cat. no. sc-18708, Santa Cruz Biotechnology, Santa Cruz, CA, USA) and Donkey-anti-Goat horseradish peroxidase (cat. no. 705-035-003, Jackson ImmunoResearch, West Grove, PA, USA). Standard ECL procedure was used for final detection (Santa Cruz Biotechnology). Localization studies were performed following transient transfection in HEK293 cells as described previously (Jansen et al., 2008).
Patch clamp electrophysiology
HEK293T cells cultured on glass coverslips were co-transfected with 0.5 μg pcDNA–KIR2.2 and 0.5 μg pEGFP1 expression constructs as described aboved. Patch clamp measurements were done using a AxoPatch 200B amplifier controlled by pClamp 9 software (Molecular devices, Sunnyvale, CA, USA). Voltage clamp measurements of whole cell IK1 were performed by applying 1 s test pulses ranging between −120 and +40 mV, in 10 mV increments, from a holding potential of −40 mV, and with series resistance compensation of at least 70%. Steady state current at the end of the pulse was normalized to cell capacitance and plotted versus test potential (corrected for liquid junction potential). Patch pipettes were made with a Sutter P-2000 puller (Sutter Instrument, Novato, CA, USA) and had resistances of 2–3 MΩ. Extracellular solution for whole cell IK1 measurements contained (in mmol/L): NaCl 140, KCl 5.4, CaCl2 1, MgCl2 1, glucose 6, NaHCO3 17.5, HEPES 15, pH 7.4/NaOH. Pipette solution contained potassium gluconate 125, KCl 10, HEPES 5, EGTA 5, MgCl2 2, CaCl2 0.6, Na2ATP 4, pH 7.20/KOH.
To evaluate the blocking effect of polyamine on KIR2.2 channel currents from both human and dog KCNJ12 genes, we analyzed chord conductance values from I–V relationship by inside-out patch clamp experiments as previously described (Ishihara and Ehara, 2004). The pipette (extracellular) solution contained (mM): 145 KCl, 1 CaCl2, and 5 HEPES (pH 7.4 with KOH). The bath (intracellular) solution contained (mM) 125 KCl, 4 EDTA (2K), 7.2 K2HPO4, and 2.8 KH2PO4 (pH 7.2 with KOH). Spermine was used in the concentration of 0.1, 1, and 10 μM. Currents were recorded from inside-out membrane patches at room temperature (22°C; De Boer et al., 2010b). The holding potential was set to 0 mV, and test pulses were applied between −60 and +90 mV in 5 mV steps with −40 mV hyperpolarizing pre-pulse. Current amplitude was measured at 2 s after the onset of the test pulse. For this evaluation, we used the equation G = I/(V − Vrev). In our study, the values of Vrev measured by a ramp protocol (40 mV/s) were always near 0 mV (between ±2 mV). Conductance values for each test voltage were normalized by the maximum value. Furthermore, we evaluated the time-dependent decay of KIR2.2 channel current just after the onset of test pulse. Single exponential fitting of the current was performed by Microcal Origin (ver.8, Microcal Software, Northampton, MA, USA).
Results and Discussion
Mapping of the KCNJ2 gene
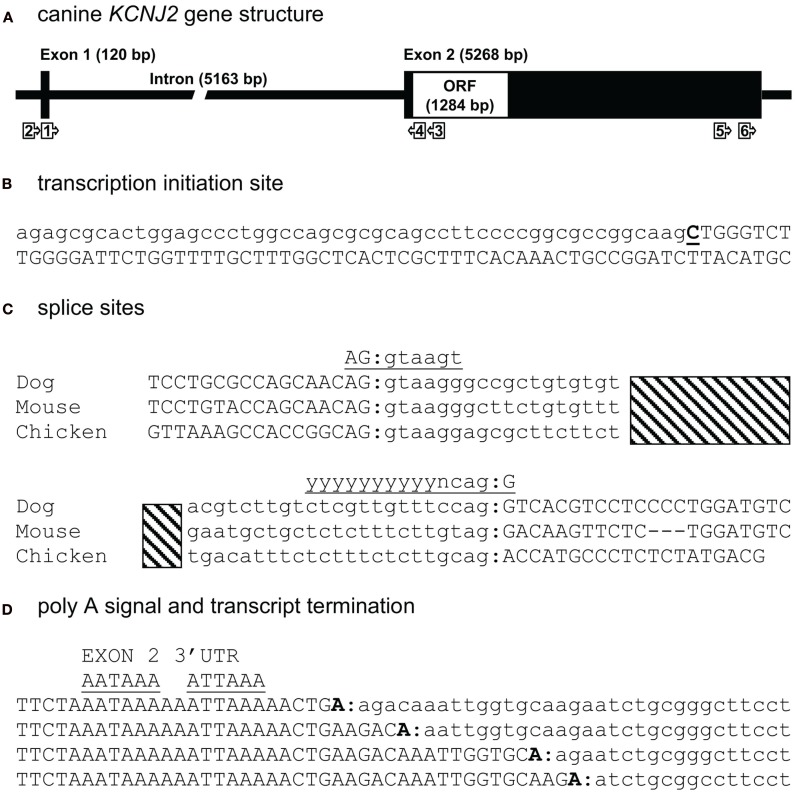
To experimentally map the canine KCNJ2 gene (Figure 1A), we first retrieved the putative KCNJ2 genomic sequence from chromosome 9 using the canine genome project data (accession number NC_006591). Homology comparison (Vector NTI suite 8, operating on Clustal W algorithm) with the mapped murine KCNJ2 (Redell and Tempel, 1998) indicated putative regions for a first and second exon. Based on these, PCR primers were designed for RT-PCR, 5′- and 3′-RACE (Table 1). RNA was isolated from canine ventricular cardiomyocytes. RT-PCR using primer KCNJ2#1se and KCNJ2#3as designed at the putative exon 1 and exon 2 sequences respectively revealed one single product of approximately 590 bp. Exon/intron boundaries were determined by sequencing of the products and aligned against the genomic DNA sequences. Boundaries were found to conform to consensus exon/intron and intron/exon sites, and high levels of homology were found at the splice sites when compared with mouse and chicken (Figure 1C). No additional exons were found in this intron region. No differences with respect to the genome reference sequence were observed.
Figure 1.
Canine KCNJ2 genomic structure. (A) Representation of the KCNJ2 gene indicating lengths of first and second exons, intron and the open reading frame (ORF). Relative position of KCNJ2 specific primers used for intron/exon mapping, 5′ and 3′ RACE are indicated by numbered box arrows. (B) Transcription initiation site (bold and underlined) with upstream (lower case) and downstream exon 1 (upper case) sequences. (C) Sequence surrounding exon 1–intron and intron–exon 2 splicing sites of dog, mouse, and chicken KCNJ2 genes. Hatched bar indicates intron region. Exon 1 and exon 2 sequences are indicated in upper case lettering, intron sequence in lower case. Consensus splicing sequences are indicated above the sites (underlined). (D) Four alternative 3′ transcript termini (bold). Exon 2 sequences are indicated in upper case, downstream genomic sequences in lower case. Consensus termination signals are indicated above the sequences (underlined).
The transcription initiation site (TIS) was mapped by 5′-RACE using primer KCNJ2#3as and the nested primer KCNJ2#4as originated from exon 2. Only a single TIS was obtained in this way (Figure 1B). RT-PCR using primer KCNJ2#2se, located directly upstream of the TIS and KCNJ2#4as yielded no product. In the mouse KCNJ2 gene, four TISs were determined located in a stretch of eight nucleotides (Redell and Tempel, 1998). In chicken, also four TISs were mapped, of which three are located adjacent to each other, while a fourth was located approximately 20 nucleotides upstream of these (Mutai et al., 2004).
For 3′-RACE, nested PCR using KCNJ2#5se and subsequent KCNJ2#6se (nested) combined with antisense primers provided by the fabricant was performed. This revealed the presence of four different transcripts (heterogeneity of mRNA cleavage) terminating after two consensus poly (A) signals, namely AAUAAA and AUUAAA (Zhao et al., 1999; Figure 1D). While for mouse, the transcript was reported to end following the first consensus poly (A) (Redell and Tempel, 1998), we found transcript termination only following the second consensus poly (A) site. Heterogenous mRNA cleavage has been noted in approximately 50% of mammalian transcripts with a single poly (A) signal (Tian et al., 2005), however its biological significance, if any, is unknown. In the case of KCNJ2, heterogenic mRNA cleavage may result from the presence of two poly (A) sites. Furthermore, it has been suggested that the RNA cleavage by the polyadenylation enzyme complex is imprecise in nature (Tian et al., 2005). Resulting sequences and annotations can be found at the following GenBank accession numbers: HM209045 for 5′ sequences, HM209046 for 3′ sequences, and BK007085 for complete gene annotation.
When comparing the overall KCNJ2 structure from dog with those of mouse and chicken (Table 2) we notice a very similar make-up existing of a short first exon (103–168 bp) containing untranslated mRNA (UTR) only, a large intron varying between ~4900 and ~5500 bp and a large second exon containing the entire ORF. While the length of the second exon is reasonably well conserved between dog and mouse (5268 and 5316 bp respectively), that of the chicken is much shorter (2020 bp). Finally, the gene structure as presented here is not annotated in NCBI Entrez Gene (GeneID 403717, accessed at July 19, 2011), that annotates the coding region only.
Table 2.
Comparison of canine KCNJ2 and KCNJ12 gene structure with that of other species.
|
KCNJ2 |
KCNJ12 |
||||
|---|---|---|---|---|---|
| Dog | Mouse1 | Chicken2 | Dog | Human3,4 | |
| Exon 1 | 120 | 168 | 103 | 406 | >192 |
| Intron 1 | 5163 | ~5500 | ~4900 | 1761 | n.d. |
| Exon 2 | 5268 | ~5316 | 2020 | 122 | ~123 |
| Intron 2 | n.a. | n.a. | n.a. | 6435 | n.d. |
| Exon 3 | n.a. | n.a. | n.a. | 1804 | ~1883 |
| ORF | 1284 | 1287 | 1284 | 1323 | 1302 |
| 5′UTR | 338 | 403 | 319 | 584 | >369 |
| 3′UTR | 3766 | ~3800 | 550 | 425 | ~527 |
Mapping of the KCNJ12 gene
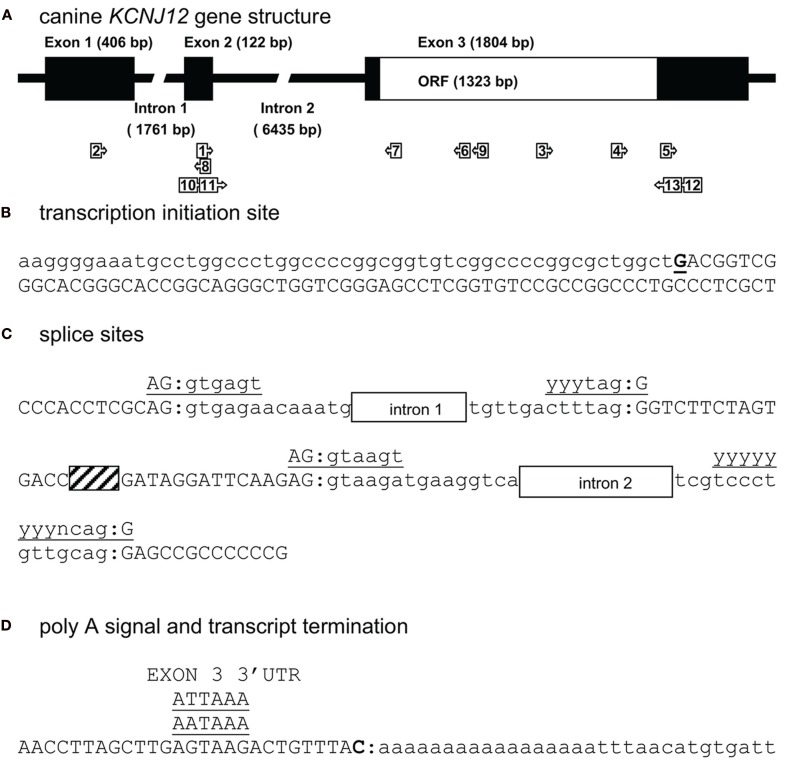
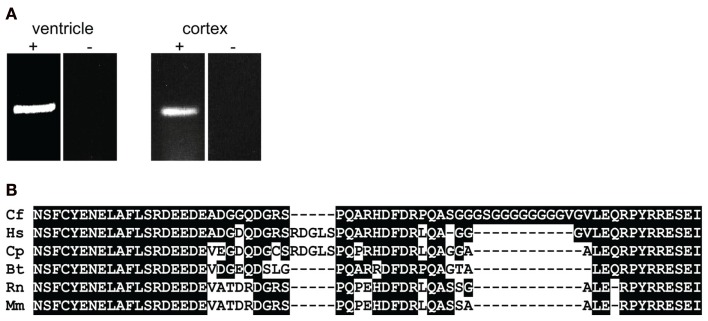
For mapping KCNJ12 (Figure 2A) experimentally, a similar approach was used as for KCNJ2. The putative KCNJ12 sequence was derived from chromosome 5 (accession number NC_006587). Next, sense (KCNJ12#1se) and antisense (KCNJ12#6as) primers were generated within exon 2 (which was then considered as the first exon) and exon 3 sequences, respectively. Subsequent sequencing revealed the exon 2 – intron 2 – exon 3 boundaries (Figure 2C). Upon 5′RACE using KCNJ12#7as located in exon 3 and the nested primer KCNJ12#8as located in exon 2, an additional exon of 406 bps was discovered (Figure 2A). Resulting sequences and annotations can be found at GenBank accession numbers HQ378597. RT-PCR using primers located in exon 1 (KCNJ12#2se) and exon 3 (KCNJ12#7as for cortex and KCNJ12#9as for right ventricle) sequences yielded a single product in heart and brain tissue (Figure 3A). Upon sequencing, the latter was found as being identical to the ventricular product. No evidence was found for alternative splicing, either by 5′RACE neither by inter-exon RT-PCR. Exon/intron boundaries were determined by sequencing PCR products and compare these to genomic DNA (Figure 2C). As for KCNJ2, boundaries were found to conform to consensus exon/intron and intron/exon sites. 5′-RACE using primer KCNJ12#7as and primer KCNJ12#8as identified one TIS (Figure 2B). No putative TATA box was found upstream of the initiation site. 3′-RACE using KCNJ12#3se and two different nested primers (KCNJ12#4se and KCNJ12#5se), all located in exon 3, identified only one termination site (Figure 2D). No genuine termination signal was identified in this gene. Finally, no differences with respect to the genome reference sequence were observed.
Figure 2.
Canine KCNJ12 genomic structure. (A) Representation of the KCNJ12 gene indicating lengths of first, second, and third exons, first and second intron, and the open reading frame (ORF). Relative position of KCNJ12 specific primers used for intron/exon mapping, 5′ and 3′ RACE are indicated by numbered box arrows 1–9. Relative positions of KCNJ12 specific primers for cloning the ORF are indicated by numbered box arrows 10–13. (B) Transcription initiation site (bold and underlined) with upstream (lower case) and downstream exon 1 (upper case) sequences. (C) Sequence surrounding exon 1–intron 1, intron 1–exon 2, exon 2–intron 2, and intron 2–exon 3 splicing sites of dog KCNJ12. Open bars indicate intron regions, hatched bar indicates exon 2 region. Exon sequences are indicated in upper case lettering, intron sequences in lower case. Consensus splicing sequences are indicated above the sites (underlined). (D) 3′ transcript terminus (bold). Exon 3 sequence is indicated in upper case, downstream genomic sequence in lower case. Consensus termination signals are indicated above the sequences (underlined).
Figure 3.
(A) KCNJ12 mRNA RT-PCR using primers located in exon 1 and 3 regions yields a single amplification product. Left panel, chronic AV block dog right ventricle; right panel control dog cortex. + Indicates prior RT-reaction, − indicates minus RT control. (B) Homology comparison of dog (Cf), human (Hs), guinea pig (Cp), bovine (Bt), rat (Rn), and mouse (Mm) KIR2.2 carboxy-termini. Conserved residues are depicted in white font on black background.
To our knowledge, no KCNJ12 gene structure from other species has been published. Indirectly however, Ryan et al. (2010) indicate that the genomic structure of KCNJ12 is very similar with that of KCNJ16. They indicate that KCNJ12 is a three exon gene with strong homology to KCNJ16 except for exon 1, which is longer in KCNJ12 than in KCNJ16. KCNJ12 intron lengths were not reported. When comparing our dog KCNJ12 sequence with that of human, strong similarity is seen for the length of exon 2 and 3 (Table 2). Furthermore, dog KCNJ12 is a three exon gene too, with its entire ORF located on Exon 3. Unfortunately, due to the lack of more specific sequence information of the human form, no reliable comparisons can be made further. Finally, the gene structure as presented here is different from the one that is annotated in NCBI Entrez Gene (GeneID 403760, accessed at July 19, 2011), that annotates the coding region on five separate exons. Furthermore, no 5′ UTR is presented, while the 3′UTR is ~22 bp in length.
Electrophysiological analysis of KIR2.2
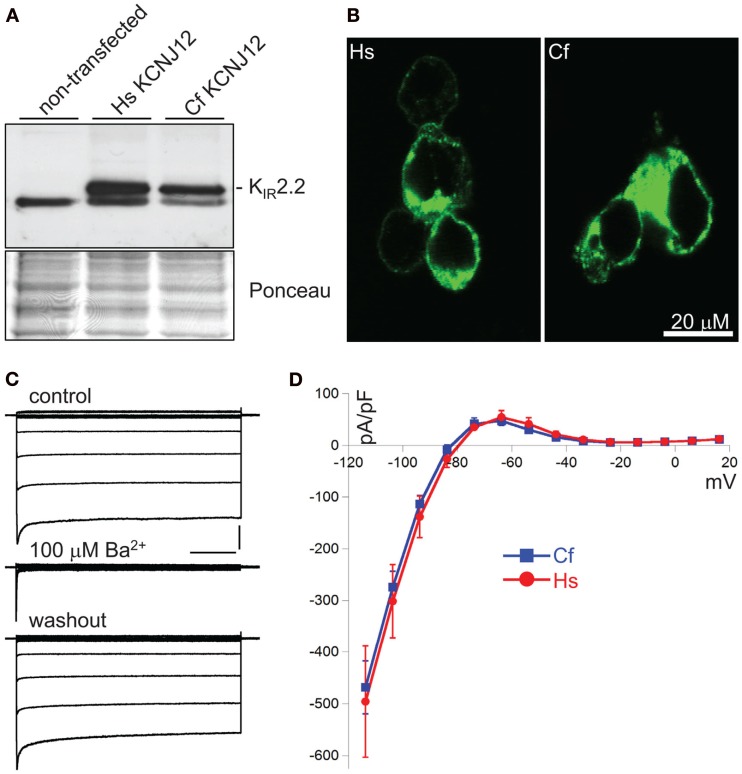
Translating genomic KCNJ12 sequence revealed a KIR2.2 protein containing a peculiar stretch of glycine residues near the carboxy-terminus. Apparently this was not a sequencing artifact as RT-PCR using primers located in exon 1 and 3 confirmed the finding in mRNA derived from left ventricle and cortex (Figure 3A). Although the glycine rich region was not identified in other species (Figure 3B), it is found in a less-conserved region of the protein. Glycine rich amino-acid stretches are found in many other proteins, however no function has been addressed. To investigate a potential functional consequence, the complete channel was cloned. Expression in COS-7 cells revealed a protein with an apparent Mw of ~50 kDa (Figure 4A). Upon transfection in HEK293 cells, strong expression of canine KIR2.2 was seen at the plasma membrane, comparable with that of human KIR2.2 (Figure 4B; Kaibara et al., 2002). When expressed in HEK293T cells, patch clamp analysis demonstrated the presence of a typical barium sensitive inward rectifying current (Figure 4C) which was indistinguishable from human KIR2.2 carried IK1 (Figure 4D).
Figure 4.
Characterization of CfKIR2.2. (A) Western blot depicting human (Hs) and dog (Cf) KIR2.2 expression upon ectopic expression in COS-7 cells. Non-transfected cells were used as negative control. (B) Subcellular localization of ectopic human (Hs) and dog (Cf) KIR2.2 in HEK293 cells. (C) Current traces of CfKIR2.2 carried IK1, before (control), upon 100 μM BaCl2 application (Ba2+), and following washout (washout). Horizontal bar: 200 ms; Vertical bar: 2 nA. (D) Current–Voltage relationship of dog (Cf, blue squares and line; n = 8, average ± SEM) and human (Hs, red circles and line; n = 4, average ± SEM) KIR2.2 carried IK1 in HEK293T cells.
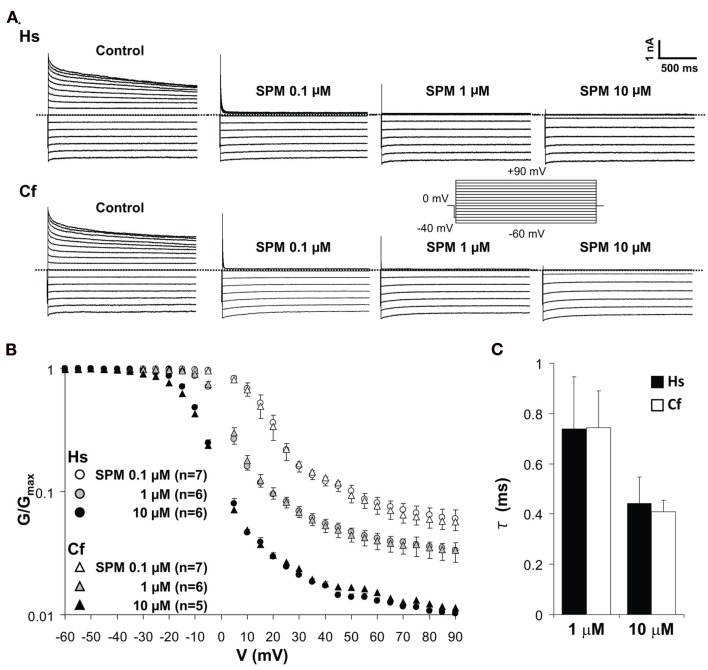
KIR2.x channels show dose-dependent changes of blocking profile by spermine (SPM) and spermidine (Ishihara and Yan, 2007). To evaluate the electrophysiological properties of canine KIR2.2 into more detail, voltage-dependent blockade of KIR2.2 channel current by different concentrations of SPM was analyzed by determining the V–G relationship (Figures 5A,B). The representative current traces measured by inside-out patch clamp show that outward IK1 currents mediated by human and dog KCNJ12 are blocked in the presence of 0.1–10 μM of SPM in a dose-dependent manner. The V–G curve shifted to more negative voltages upon increasing the SPM concentration. No significant difference in V–G relations at either SPM concentration was observed (Figure 5B). In the presence of physiological levels of SPM (1 and 10 μM; Yan et al., 2005), human and dog KIR2.2 channels displayed similar time-dependent decay at +80 mV [1 μM: 0.74 ± 0.21 (SEM; n = 5) and 0.74 ± 0.15 (n = 6) ms; 10 μM: 0.44 ± 0.10 (n = 5) and 0.41 ± 0.06 (n = 5) ms for human and dog respectively; Figure 5C]. Our results suggested that there is no difference between human and dog KIR2.2 with respect to SPM affinity and speed of binding. As the glycine rich domain is not conserved among species and we observed no differences in electrophysiological parameters tested, we speculate that it has no relevance for canine KIR2.2 function and its interaction with polyamines.
Figure 5.
Spermine-induced block of human (Hs) and dog (Cf) KIR2.2 channels. (A) Representative traces of KIR2.2 channel currents in the absence and presence of 0.1, 1, and 10 μM of spermine. Inset: pulse protocol, currents were elicit by 10 mV step test pulses from −60 to +90 mV. (B) Comparison of V–G relationships of human and dog KIR2.2. The scale of G/Gmax is logarithmic. Test pulses were applied in 5 mV steps from −60 to +90 mV, and G was calculated for each voltage. (C) The comparison of time constant of KIR2.2 channel currents at +80 mV in the presence of 1 and 10 μM SPM.
Only few studies have addressed KIR2.2 in ventricular tissue of large animal models, and currently, we can only speculate on the function and significance of canine KIR2.2 expression in the heart. Ojaimi et al. (2007) measured expression of left ventricular genes upon transition to pacing induced heart failure in the dog heart. Their data indicate clear expression of both KCNJ2 and KCNJ12 in the left ventricle (NCBI gene expression omnibus (GEO) profiles and GDS2424/1582618_at and GDS2424/1582977_at, respectively), however, no change was found upon transition into heart failure. Zobel et al. (2003) report KIR2.2 expression in isolated rabbit ventricular myocytes. Upon culturing, expression levels remained whereas KIR2.1 expression decreased by 80%. In humans, KIR2.2 displayed a more pronounced regional expression than KIR2.1, i.e., a preferential expression in the epicardium of the right ventricle compared to the endocardium (Gaborit et al., 2007).
Conclusion
We were able to experimentally map the canine KCNJ2 and KCNJ12 genes. KCNJ2 is a two exon gene that displays a similar organization as the mouse homolog, while it differs with the chicken variant in the length of exon 2 only. KCNJ12 is a three exon gene, which resembles the human organization. No alternative splicing in KCNJ12 was detected in heart or brain tissue. Canine KCNJ12 produced genuine KIR2.2 carried IK1 upon expression in HEK293T cells.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
Dr. H. Takanari was supported during this work through the SENSHIN Medical Research Foundation.
References
- Anumonwo J. M., Lopatin A. N. (2010). Cardiac strong inward rectifier potassium channels. J. Mol. Cell. Cardiol. 48, 45–54 10.1016/j.yjmcc.2009.08.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coronel R. (2010). Challenging cardiac electrophysiology. Front. Physiol. 1:8. 10.3389/fphys.2010.00008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Boer T. P., Houtman M. J. C., Compier M., Van der Heyden M. A. G. (2010a). The mammalian KIR2.x inward rectifier ion channel family: expression pattern and pathophysiology. Acta Physiol. 199, 243–253 [DOI] [PubMed] [Google Scholar]
- De Boer T. P., Nalos L., Stary A., Kok B., Houtman M. J. C., Antoons G., Van Veen T. A. B., Beekman J. D. M., De Groot B. L., Opthof T., Rook M. B., Vos M. A., Van der Heyden M. A. G. (2010b). The anti-protozoal drug pentamidine blocks KIR2.x-mediated inward rectifier current by entering the cytoplasmic pore region of the channel. Br. J. Pharmacol. 159, 1532–1541 10.1111/j.1476-5381.2010.00658.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhamoon A. S., Jalife J. (2005). The inward rectifier current (IK1) controls cardiac excitability and is involved in arrhythmogenesis. Heart Rhythm 2, 316–324 10.1016/j.hrthm.2004.11.012 [DOI] [PubMed] [Google Scholar]
- Flicek P. (2007). Gene prediction: compare and CONTRAST. Genome Biol. 8, 233. 10.1186/gb-2007-8-12-233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaborit N., Le Bouter S., Szuts V., Varro A., Escande D., Nattel S., Demolombe S. (2007). Regional and tissue specific transcript signatures of ion channel genes in the non-diseased human heart. J. Physiol. 582, 675–693 10.1113/jphysiol.2006.126714 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hugnot J. P., Pedeutour F., Le Calvez C., Grosgeorge J., Passage E., Fontes M., Lazdunski M. (1997). The human inward rectifying K+ channel Kir2.2 (KCNJ12) gene: gene structure, assignment to chromosome 17p11.1, and identification of a simple tandem repeat polymorphism. Genomics 39, 113–116 10.1006/geno.1996.4450 [DOI] [PubMed] [Google Scholar]
- Ishihara K., Ehara T. (2004). Two modes of polyamine block regulating the cardiac inward rectifier K+ current IK1 as revealed by a study of the Kir2.1 channel expressed in a human cell line. J. Physiol. 556, 61–78 10.1113/jphysiol.2003.055434 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ishihara K., Yan D. H. (2007). Low-affinity spermine block mediating outward currents through Kir2.1 and Kir2.2 inward rectifier potassium channels. J. Physiol. 583, 891–908 10.1113/jphysiol.2007.136028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jansen J. A., De Boer T. P., Wolswinkel R., Van Veen T. A., Vos M. A., Van Rijen H. V. M., Van der Heyden M. A. G. (2008). Lysosome mediated Kir2.1 breakdown directly influences inward rectifier current density. Biochem. Biophys. Res. Commun. 367, 687–692 10.1016/j.bbrc.2007.12.168 [DOI] [PubMed] [Google Scholar]
- Kaibara M., Ishihara K., Doi Y., Hayashi H., Ehara T., Taniyama K. (2002). Identification of human Kir2.2 (KCNJ12) gene encoding functional inward rectifier potassium channel in both mammalian cells and Xenopus oocytes. FEBS Lett. 531, 250–254 10.1016/S0014-5793(02)03512-3 [DOI] [PubMed] [Google Scholar]
- Li J., McLerie M., Lopatin A. (2004). Transgenic upregulation of IK1 in the mouse heart leads to multiple abnormalities of cardiac excitability. Am. J. Physiol. Heart Circ. Physiol. 287, H2790–H2802 10.1152/ajpheart.00124.2004 [DOI] [PubMed] [Google Scholar]
- Lindblad-Toh K., Wade C. M., Mikkelsen T. S., Karlsson E. K., Jaffe D. B., Kamal M., Clamp M., Chang J. L., Kulbokas E. J., III, Zody M. C., Mauceli E., Xie X., Breen M., Wayne R. K., Ostrander E. A., Ponting C. P., Galibert F., Smith D. R., DeJong P. J., Kirkness E., Alvarez P., Biagi T., Brockman W., Butler J., Chin C. W., Cook A., Cuff J., Daly M. J., DeCaprio D., Gnerre S., Grabherr M., Kellis M., Kleber M., Bardeleben C., Goodstadt L., Heger A., Hitte C., Kim L., Koepfli K. P., Parker H. G., Pollinger J. P., Searle S. M., Sutter N. B., Thomas R., Webber C., Baldwin J., Abebe A., Abouelleil A., Aftuck L., Ait-Zahra M., Aldredge T., Allen N., An P., Anderson S., Antoine C., Arachchi H., Aslam A., Ayotte L., Bachantsang P., Barry A., Bayul T., Benamara M., Berlin A., Bessette D., Blitshteyn B., Bloom T., Blye J., Boguslavskiy L., Bonnet C., Boukhgalter B., Brown A., Cahill P., Calixte N., Camarata J., Cheshatsang Y., Chu J., Citroen M., Collymore A., Cooke P., Dawoe T., Daza R., Decktor K., DeGray S., Dhargay N., Dooley K., Dooley K., Dorje P., Dorjee K., Dorris L., Duffey N., Dupes A., Egbiremolen O., Elong R., Falk J., Farina A., Faro S., Ferguson D., Ferreira P., Fisher S., FitzGerald M., Foley K., Foley C., Franke A., Friedrich D., Gage D., Garber M., Gearin G., Giannoukos G., Goode T., Goyette A., Graham J., Grandbois E., Gyaltsen K., Hafez N., Hagopian D., Hagos B., Hall J., Healy C., Hegarty R., Honan T., Horn A., Houde N., Hughes L., Hunnicutt L., Husby M., Jester B., Jones C., Kamat A., Kanga B., Kells C., Khazanovich D., Kieu A. C., Kisner P., Kumar M., Lance K., Landers T., Lara M., Lee W., Leger J. P., Lennon N., Leuper L., LeVine S., Liu J., Liu X., Lokyitsang Y., Lokyitsang T., Lui A., Macdonald J., Major J., Marabella R., Maru K., Matthews C., McDonough S., Mehta T., Meldrim J., Melnikov A., Meneus L., Mihalev A., Mihova T., Miller K., Mittelman R., Mlenga V., Mulrain L., Munson G., Navidi A., Naylor J., Nguyen T., Nguyen N., Nguyen C., Nguyen T., Nicol R., Norbu N., Norbu C., Novod N., Nyima T., Olandt P., O’Neill B., O’Neill K., Osman S., Oyono L., Patti C., Perrin D., Phunkhang P., Pierre F., Priest M., Rachupka A., Raghuraman S., Rameau R., Ray V., Raymond C., Rege F., Rise C., Rogers J., Rogov P., Sahalie J., Settipalli S., Sharpe T., Shea T., Sheehan M., Sherpa N., Shi J., Shih D., Sloan J., Smith C., Sparrow T., Stalker J., Stange-Thomann N., Stavropoulos S., Stone C., Stone S., Sykes S., Tchuinga P., Tenzing P., Tesfaye S., Thoulutsang D., Thoulutsang Y., Topham K., Topping I., Tsamla T., Vassiliev H., Venkataraman V., Vo A., Wangchuk T., Wangdi T., Weiand M., Wilkinson J., Wilson A., Yadav S., Yang S., Yang X., Young G., Yu Q., Zainoun J., Zembek L., Zimmer A., Lander E. S. (2005). Genome sequence, comparative analysis and haplotype structure of the domestic dog. Nature 438, 803–819 10.1038/nature04338 [DOI] [PubMed] [Google Scholar]
- Michael G., Xiao L., Qi X. Y., Dobrev D., Nattel S. (2009). Remodelling of cardiac repolarization: how homeostatic responses can lead to arrhythmogenesis. Cardiovasc. Res. 81, 491–499 10.1093/cvr/cvn266 [DOI] [PubMed] [Google Scholar]
- Mutai H., Kenyon L. C., Locke E., Kikuchi N., Oberholtzer J. C. (2004). Characterization of the chicken inward rectifier K+-channel IRK1/Kir2.1 gene. BMC Genomics 5, 90. 10.1186/1471-2164-5-90 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojaimi C., Qanud K., Hintze T. H., Recchia F. A. (2007). Altered expression of a limited number of genes contributes to cardiac decompensation during chronic ventricular tachypacing in dogs. Physiol. Genomics 29, 76–83 [DOI] [PubMed] [Google Scholar]
- Oros A., Beekman J. D., Vos M. A. (2008). The canine model with chronic, complete atrio-ventricular block. Pharmacol. Ther. 119, 168–178 10.1016/j.pharmthera.2008.03.006 [DOI] [PubMed] [Google Scholar]
- Priori S. G., Pandit S. V., Rivolta I., Berenfeld O., Ronchetti E., Dhamoon A., Napolitano C., Anumonwo J., Di Barletta M. R., Gudapakkam S., Bosi G., Stramba-Badiale M., Jalife J. (2005). A novel form of short QT syndrome (SQT3) is caused by a mutation in the KCNJ2 gene. Circ. Res. 96, 800–807 10.1161/01.RES.0000162101.76263.8c [DOI] [PubMed] [Google Scholar]
- Redell J. B., Tempel B. L. (1998). Multiple promoter elements interact to control transcription of the potassium channel gene, KCNJ2. J. Biol. Chem. 273, 22807–22818 10.1074/jbc.273.35.22807 [DOI] [PubMed] [Google Scholar]
- Roden D. M. (1998). Taking the “idio” out of “idiosyncratic”: predicting torsades de pointes. Pacing Clin. Electrophysiol. 21, 1029–1034 10.1111/j.1540-8159.1998.tb00148.x [DOI] [PubMed] [Google Scholar]
- Rodríguez-Menchaca A. A., Navarro-Polanco R. A., Ferrer-Villada T., Rupp J., Sachse F. B., Tristani-Firouzi M., Sánchez-Chapula J. A. (2008). The molecular basis of chloroquine block of the inward rectifier KIR2.1 channel. Proc. Natl. Acad. Sci. U.S.A. 105, 1364–1368 10.1073/pnas.0708153105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ryan D. P., Da Silva M. R., Soong T. W., Fontaine B., Donaldson M. R., Kung A. W., Jongjaroenprasert W., Liang M. C., Khoo D. H., Cheah J. S., Ho S. C., Bernstein H. S., Maciel R. M., Brown R. H., Jr., Ptácek L. J. (2010). Mutations in potassium channel Kir2.6 cause susceptibility to thyrotoxic hypokalemic periodic paralysis. Cell 140, 88–98 10.1016/j.cell.2009.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thomsen M. B., Matz J., Volders P. G., Vos M. A. (2006a). Assessing the proarrhythmic potential of drugs: current status of models and surrogate parameters of torsades de pointes arrhythmias. Pharmacol. Ther. 112, 150–170 10.1016/j.pharmthera.2005.04.009 [DOI] [PubMed] [Google Scholar]
- Thomsen M. B., Volders P. G., Beekman J. D., Matz J., Vos M. A. (2006b). Beat-to-beat variability of repolarization determines proarrhythmic outcome in dogs susceptible to drug-induced torsades de pointes. J. Am. Coll. Cardiol. 48, 1268–1276 10.1016/j.jacc.2006.05.048 [DOI] [PubMed] [Google Scholar]
- Thomsen M. B., Oros A., Schoenmakers M., Van Opstal J. M., Maas J. N., Beekman J. D., Vos M. A. (2007). Proarrhythmic electrical remodelling is associated with increased beat-to-beat variability of repolarisation. Cardiovasc. Res. 73, 521–530 10.1016/j.cardiores.2006.11.025 [DOI] [PubMed] [Google Scholar]
- Tian B., Hu J., Zhang H., Lutz C. S. (2005). A large-scale analysis of mRNA polyadenylation of human and mouse genes. Nucleic Acids Res. 33, 201–212 10.1093/nar/gki158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tristani-Firouzi M., Etheridge S. P. (2010). Kir 2.1 channelopathies: the Andersen-Tawil syndrome. Pflugers Arch. 460, 289–294 10.1007/s00424-010-0820-6 [DOI] [PubMed] [Google Scholar]
- Vega A. L., Tester D. J., Ackerman M. J., Makielski J. C. (2009). Protein kinase A-dependent biophysical phenotype for V227F-KCNJ2 mutation in catecholaminergic polymorphic ventricular tachycardia. Circ. Arrhythm. Electrophysiol. 2, 540–547 10.1161/CIRCEP.109.872309 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xia M., Jin Q., Bendahhou S., He Y., Larroque M. M., Chen Y., Zhou Q., Yang Y., Liu Y., Liu B., Zhu Q., Zhou Y., Lin J., Liang B., Li L., Dong X., Pan Z., Wang R., Wan H., Qiu W., Xu W., Eurlings P., Barhanin J., Chen Y. (2005). A Kir2.1 gain-of-function mutation underlies familial atrial fibrillation. Biochem. Biophys. Res. Commun. 332, 1012–1019 10.1016/j.bbrc.2005.05.054 [DOI] [PubMed] [Google Scholar]
- Yan D. H., Nishimura K., Yoshida K., Nakahira K., Ehara T., Igarashi K., Ishihara K. (2005). Different intracellular polyamine concentrations underlie the difference in the inward rectifier K+ currents in atria and ventricles of the guinea pig heart. J. Physiol. 563, 713–724 10.1113/jphysiol.2004.077677 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Lin H., Xiao J., Lu Y., Luo X., Li B., Zhang Y., Xu C., Bai Y., Wang H., Chen G., Wang Z. (2007). The muscle-specific microRNA miR-1 regulates cardiac arrhythmogenic potential by targeting GJA1 and KCNJ2. Nat. Med. 13, 486–491 10.1038/nm1547 [DOI] [PubMed] [Google Scholar]
- Zaritsky J. J., Redell J. B., Tempel B. L., Schwarz T. L. (2001). The consequences of disrupting cardiac inwardly rectifying K+ current (IK1) as revealed by the targeted deletion of the murine Kir2.1 and Kir2.2 genes. J. Physiol. 533, 697–710 10.1111/j.1469-7793.2001.t01-1-00697.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao J., Hyman L., Moore C. (1999). Formation of mRNA 3′ ends in eukaryotes: mechanism, regulation, and interrelationships with other steps in mRNA synthesis. Microbiol. Mol. Biol. Rev. 63, 405–445 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zobel C., Cho H. C., Nguyen T. T., Pekhletski R., Diaz R. J., Wilson G. J., Backx P. H. (2003). Molecular dissection of the inward rectifier potassium current (IK1) in rabbit cardiomyocytes: evidence for heteromeric co-assembly of Kir2.1 and Kir2.2. J. Physiol. 550, 365–372 10.1113/jphysiol.2002.036400 [DOI] [PMC free article] [PubMed] [Google Scholar]