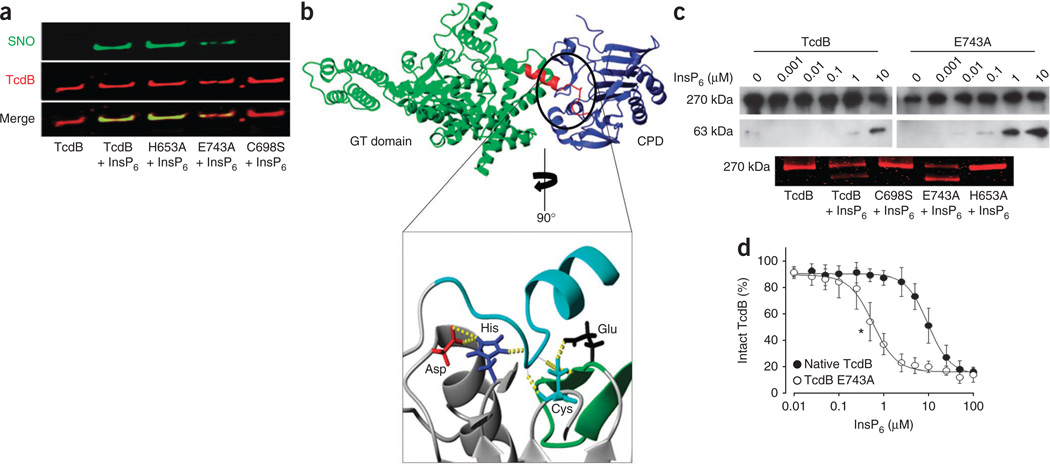
Figure 3.
A catalytic-site motif for S-nitrosylation. (a) Effect of InsP6 on S-nitrosylation of the TcdB catalytic site and TcdB catalytic-site mutants (His653Ala, Glu743Ala and Cys698Ser). GSNO (100 µM) and InsP6 (100 µM) were added for 10 min at 37 °C. (b) Extensive hydrogen bonding within the active site of the N-terminus RTXVC CPD11 (inset, yellow dotted lines) interconnects a catalytic-site S-nitrosylation motif that is conserved in TcdA and TcdB (Supplementary Figs. 7 and 14). The substrate domain (autocleavage site) is shown in turquoise (inset) and in red (top), where it bridges TcdB glucosyltransferase (GT) and cysteine protease (blue) domains, as shown in the combined crystal structure model, described further in Supplementary Figure 13. (c) Allosteric effect of InsP6 on catalytic activity of native TcdB versus catalytic-site mutants. Top, immunoblot showing autocleavage of native TcdB versus Glu743Ala mutant in the presence of InsP6. Bottom, GelCode Blue staining showing allosteric influence of InsP6 on catalytic activity of native TcdB versus catalytic-site mutants (C698S, E743A and H653A). (d) Autocleavage of native TcdB versus E743A mutant with increasing InsP6 concentrations (n = 3; error bars show s.e.m. *P < 001 for the lowest significant InsP6 concentration, Mann-Whitney U test for ranks).