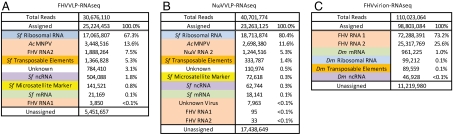
Fig. 2.
Summaries of the alignments of reads from each dataset, grouped into functional RNA classes. (A) FHVVLP-RNAseq, (B) NωVVLP-RNAseq, and (C) FHVvirion-RNAseq. Color-coded: peach, viral RNA; green, mRNA; blue, rRNA; gold, transposon RNA; purple, ncRNA; and yellow, microsatellite markers from ref. 21. In A and B, assembled contigs that did not have significant homology to other known sequences are indicated as unknown. Dm, Drosophila melanogaster; FHV, flock house virus; Sf, Spodoptera frugiperda; AcMNPV, Autographa californica multiple nuclear polyhedrosis virus.