Figure 1.
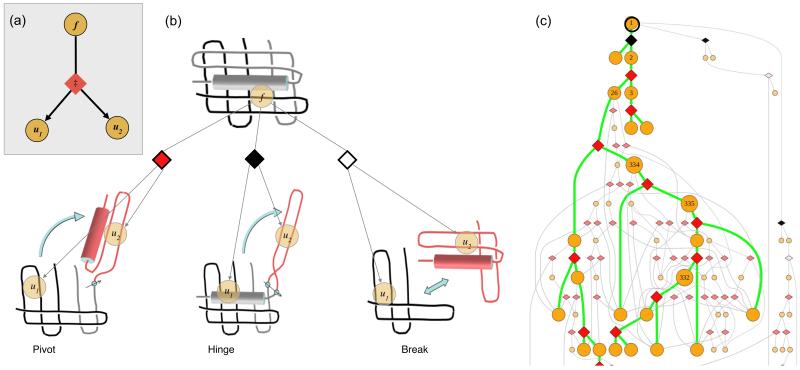
(a) Elemental subsystem for the kinetic model, a cut. f is any spatially contiguous substructure of a protein, and is partitioned into spatially contiguous substructures u1 and u2. (b) Diamond shapes represent the cuts, each having a type (color) and an associated energy barrier ‡. A pivot motion is single point revolute joint. A hinge rotates around two points. A break is a pure translational motion. Rotations and translations must not cross chains. (c) Top portion of an unfolding pathway DAG for DHFR. Node 1 is the fully folded state. Green lines indicate the pathway of maximum unfolding traffic; grey lines are other significant pathways. Unfolding simulations start with all of the protein in node 1, and end when node concentrations reach equilibrium.