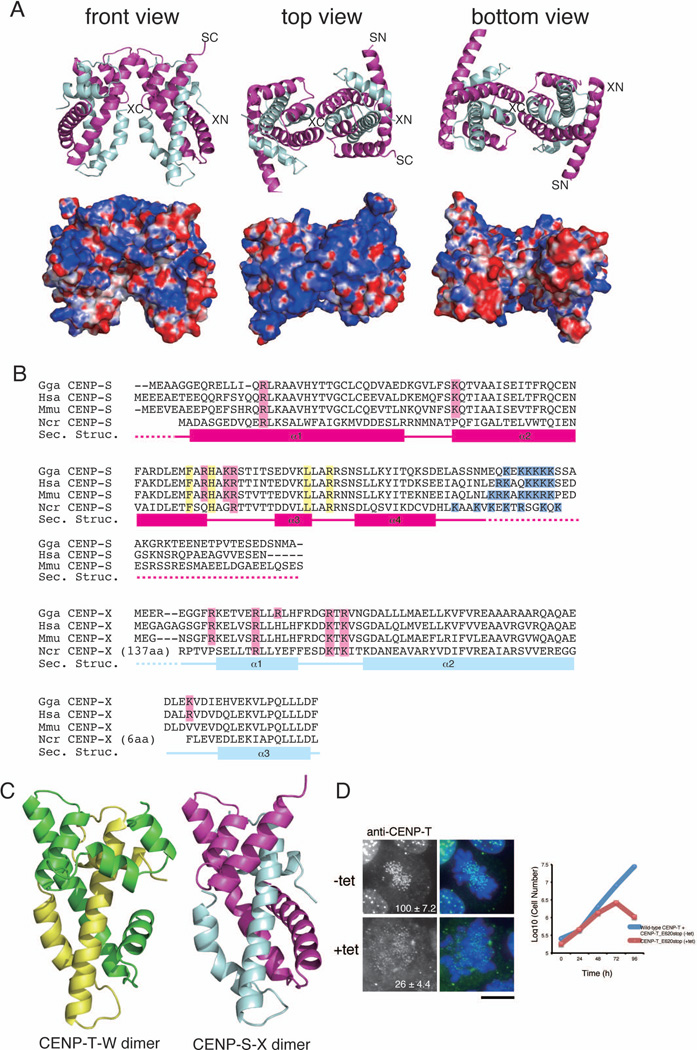
Figure 2. (refers to Supplemental Figure S2 and Table S2). The CENP-S-X complex forms a tetramer with structural similarity to the CENP-T-W complex.
(A) Ribbon diagram representations of the (CENP-S-X)2 tetramer (top) and its surface charges (bottom). Three orthogonal views of the (CENP-S-X)2 tetramer are shown. CENP-S is colored in magenta and CENP-X is colored in cyan. The N- and C-terminus of CENP-S and CENP-X are shown as SN, SC, XN, and XC, respectively. Electrostatic surface charges of CENP-S and CENP-X were calculated by APBS and are contoured from −8.0 (red) to 8.0 (blue).
(B) Sequence alignment of CENP-S (top) and CENP-X (bottom) from chicken, human, mouse, and Neurospora. Boxes (α1 – α4), solid lines, and dashed lines indicate α-helices, random coil regions, and disordered residues, respectively. Residues marked by pink are predicted as DNA binding sites by comparison with canonical histones and residues marked by yellow indicate the tetramerization interface. Basic residues marked by blue are additional DNA binding sites of CENP-S.
(C) Structural comparison of the CENP-T-W and CENP-S-X complexes. Both complexes are viewed from the same angle as in (A).
(D) Immunofluorescence analysis of DT40 cells in which endogenous CENP-T is replaced by the CENP-TE620stop mutant with deletion of the α5 helix (+tet). In the absence of tetracycline (−tet), cells express both wild-type CENP-T and the CENP-TE620stop mutant. In the presence of tetracycline (+tet), cells express only CENP-TE620stop mutant. The growth curve was shown. Bar, 10 µm.