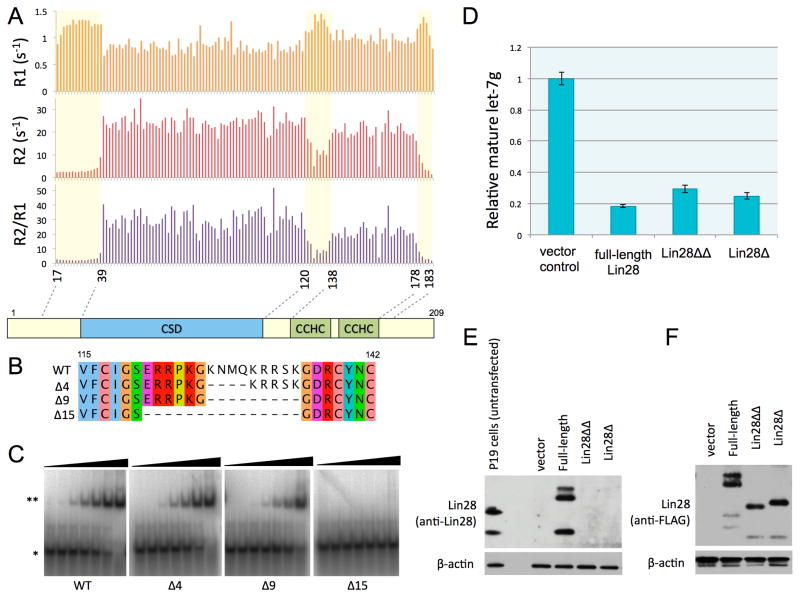
Figure 2.
Linker between CSD and CCHCx2 is flexible. See also Figure S2. (A) Longitudinal (R1) and transverse (R2) relaxation rates and the ratio (R2/R1), plotted against the residue number. Relatively more dynamic regions are marked with a light yellow box. (B) Alignment of internal deletions in the linker, indicated with the number of amino acids deleted on left. (C) EMSAs with preE-let-7d as probe, mixed with increasing concentrations (0.005, 0.02, 0.08, 0.3, 1.2, 5, 20μM) of linker deletion constructs of Lin28(16–184): *, free probe; **, complex. (D) Quantitative RT-PCR results for in vivo levels of mature let-7g. Lin28Δ is truncated at both N and C termini. Lin28ΔΔ has both of the terminal extensions and the linker removed. The standard deviation is calculated from triplicate experiments; U6 RNA levels were used for normalization. (E) and (F) Western blots of Trizol bottom layer for transfections shown in (D). Anti-Lin28 antibodies do not recognize truncation constructs, so anti-FLAG was used in (F) to compare the relative expression levels of different Lin28 constructs.