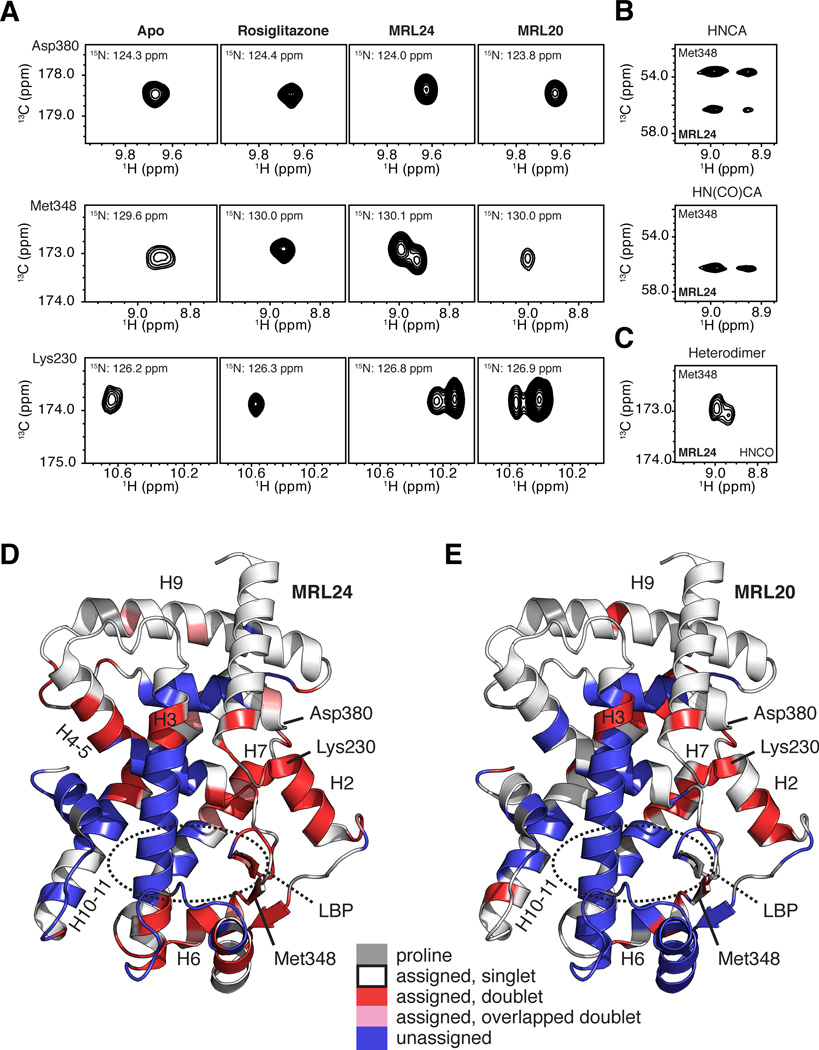
Figure 3. NMR data implicates slow conformational exchange between two receptor conformations in the mechanism of action of graded PPARγ modulators.
(A) Regions of 3D TROSY-HNCO spectra for PPARγ LBD in the apo form or bound to rosiglitazone, MRL24 and MRL20. NMR resonances for Asp380, Met348 and Lys230 are fully stabilized when bound to rosiglitazone, but display varying degrees of two resonance populations when bound to MRL24 and MRL20. (B–C) The two resonance populations were present in (B) data used for backbone assignments, including 3D TROSY-HNCA and TROSY-HN(CO)CA, and (C) in 3D TROSY-HNCO data for [2H,13C,15N]-PPARγ/[unlabeled]-RXRα LBD heterodimer bound to MRL24 and 9-cis-retinoic acid, respectively. (D,E) PPARγ LBD structure (PDB 1FM6) colored by residues with two chemical shift populations (red), line shapes suggesting two populations that have significant overlap (pink), or with missing assignments likely from intermediate conformational exchange (blue) in the presence of MRL24. Proline residues are colored grey. See also Figures S1 and S2.