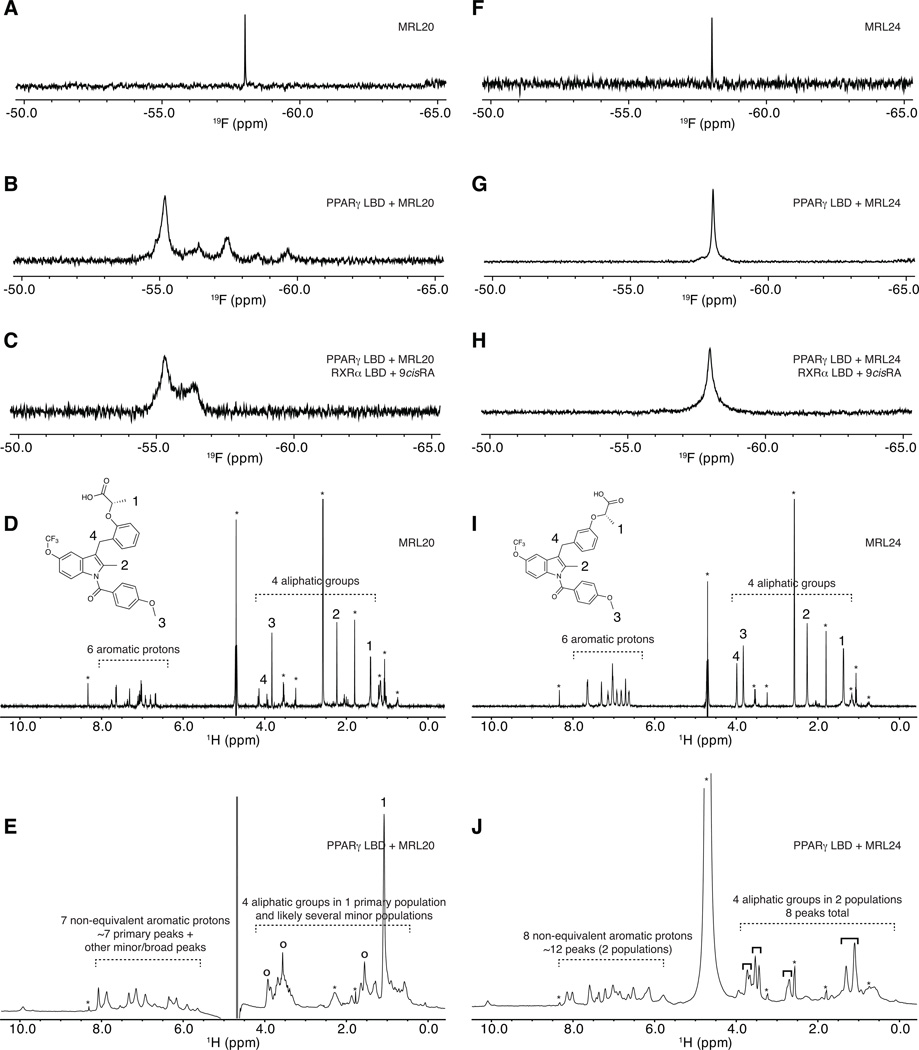
Figure 4. Receptor-bound ligand-observe NMR data reveals multiple binding modes for graded PPARγ modulators.
Ligand-observe 1D NMR data for MRL20 (A–E) and MRL24 (F–J), including 19F (A–C,F–H) and 1H (D,E,I,J) NMR data. 19F NMR data for MRL20 reveals that the single CF3 group (A) displays multiple populations when bound to PPARγ (B) and the PPARγ/RXRα heterodimer (C), including one primary state and one-to-several lowly populated states. 1H NMR data for MRL20 also reveals the aromatic and aliphatic groups (D) display a similar profile when bound to PPARγ (E). 19F NMR data for the single CF3 group of MRL24 (F) reveals a single resonance population when bound to PPARγ (G) or the PPARγ/RXRα heterodimer (H). 1H NMR data for MRL24 reveals that the aromatic and aliphatic NMR resonances (I) are in two populations when bound to PPARγ (J). Buffer signals are marked with an asterisk in (D,E,I,J); primary MRL20 signals are marked with open circles in (E); four groups of two MRL24 populations are marked with solid black brackets in (J). Figure S3 shows a zoomed in view of the aliphatic region for MRL24 in (J).