Abstract
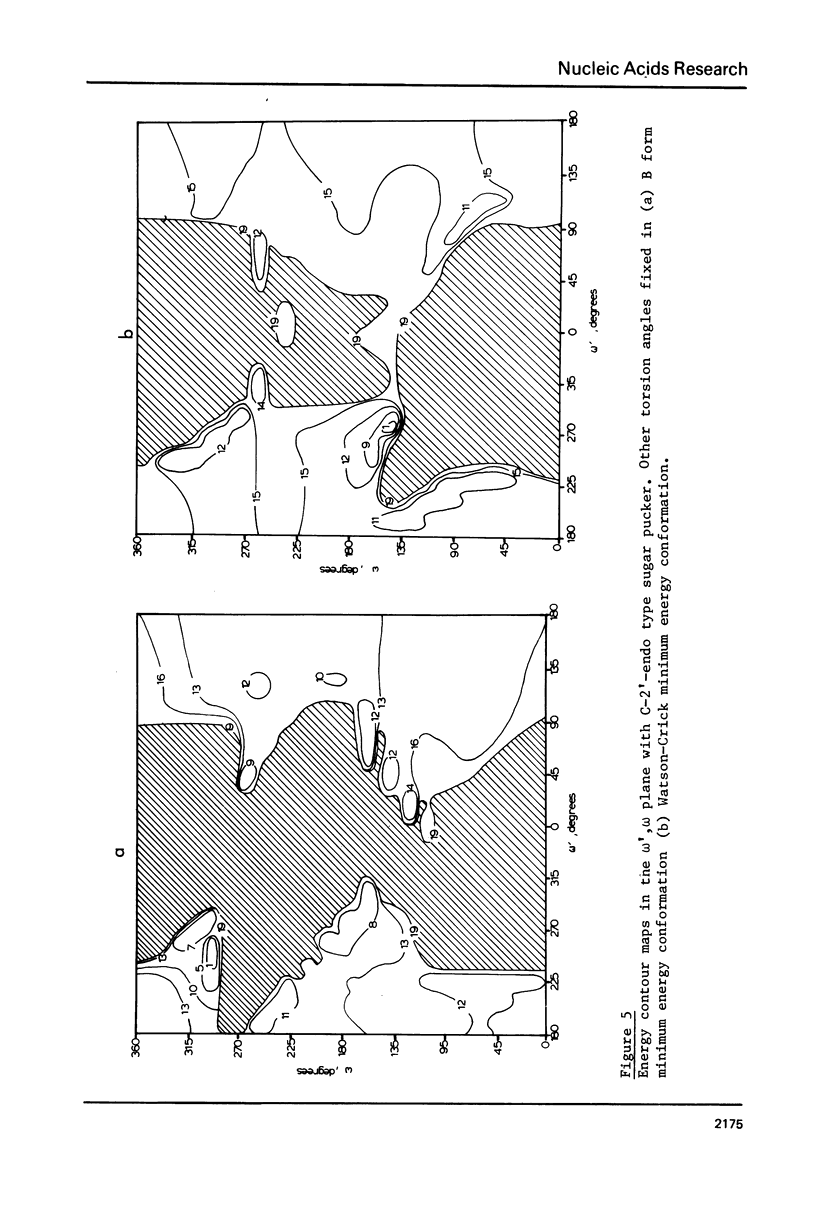
Addition of 3' and 5' terminal phosphates to dApdA causes a decrease in conformational flexibility. pdApdAp has much fewer conformers with energies below 2.5 kcal./mole than dApdA. THE A, B and Watson-Crick (34) helices are the most preferred forms. Other important conformations are in the trans domain of psi. Thus, flexibility in psi as well as in omega and omega, and in the sugar pucker is indicated. The transformation from the B helix to the Watson-Crick helix follows a low energy path. This is significant since Watson-Crick conformations may be important for intercalation into nucleic acid polymers (40-42) above the dimer level. The B helix is preferred over the A form in these large DNA subunits.
Full text
PDF








Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Alden C. J., Arnott S. Stereochemical model for proflavin intercalation in A-DNA. Nucleic Acids Res. 1977 Nov;4(11):3855–3861. doi: 10.1093/nar/4.11.3855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alden C. J., Arnott S. Visualization of planar drug intercalations in B-DNA. Nucleic Acids Res. 1975 Oct;2(10):1701–1717. doi: 10.1093/nar/2.10.1701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altona C., Sundaralingam M. Conformational analysis of the sugar ring in nucleosides and nucleotides. A new description using the concept of pseudorotation. J Am Chem Soc. 1972 Nov 15;94(23):8205–8212. doi: 10.1021/ja00778a043. [DOI] [PubMed] [Google Scholar]
- Arnott S., Chandrasekaran R., Leslie A. G. Structure of the single-stranded polyribonucleotide polycytidylic acid. J Mol Biol. 1976 Sep 25;106(3):735–748. doi: 10.1016/0022-2836(76)90262-x. [DOI] [PubMed] [Google Scholar]
- Arnott S., Hukins D. W. Conservation of conformation in mono and poly-nucleotides. Nature. 1969 Nov 29;224(5222):886–888. doi: 10.1038/224886a0. [DOI] [PubMed] [Google Scholar]
- Broyde S. B., Stellman S. D., Hingerty B., Langridge R. Conformational stability in dinucleoside phosphate crystals. Semiempirical potential energy calculations for uridylyl-3'-5'-adenosine monophosphate (UpA) and guanylyl-3',5'-cytidine monophosphate (GpC). Biopolymers. 1974 Jun;13(6):1243–1259. doi: 10.1002/bip.1974.360130615. [DOI] [PubMed] [Google Scholar]
- Broyde S. B., Wartell R. M., Stellman S. D., Hingerty B., Langridge R. Classical potential energy calculations for ApA, CpC, GpG, and UpU. The influence of the bases on RNA subunit conformations. Biopolymers. 1975 Aug;14(8):1597–1613. doi: 10.1002/bip.1975.360140805. [DOI] [PubMed] [Google Scholar]
- Broyde S., Hingerty B. 'A' forms of RNAs in single strands, duplexes and RNA-DNA hybrids. Nucleic Acids Res. 1978 Aug;5(8):2729–2741. doi: 10.1093/nar/5.8.2729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D. M., Dhingra M. M., Sarma R. H. Spatial configuration of deoxyribotrinucleoside diphosphates in aqueous solution. Nucleic Acids Res. 1978 Nov;5(11):4399–4416. doi: 10.1093/nar/5.11.4399. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng D. M., Sarma R. H. Intimate details of the conformational characteristics of deoxyribodinucleoside monophosphates in aqueous solution. J Am Chem Soc. 1977 Oct 26;99(22):7333–7348. doi: 10.1021/ja00464a038. [DOI] [PubMed] [Google Scholar]
- Fujii S., Tomita K. Conformational analysis of polynucleotides. I. The favorable left-handed helical model for the poly(8,2'-S-cycloadenylic acid) with high anti conformation. Nucleic Acids Res. 1976 Aug;3(8):1973–1984. doi: 10.1093/nar/3.8.1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Govil G. Conformational structure of polynucleotides around the O-P bonds: refined parameters for CPF calculations. Biopolymers. 1976 Nov;15(11):2303–2307. doi: 10.1002/bip.1976.360151119. [DOI] [PubMed] [Google Scholar]
- Govil G., Fisk C., Howard F. B., Miles H. T. Structure of poly 8-bromoadenylic acid; conformational studies by CPF energy calculations. Nucleic Acids Res. 1977 Aug;4(8):2573–2592. doi: 10.1093/nar/4.8.2573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingerty B., Broyde S. A model for the single stranded random coil form of polydeoxyadenylic acid from minimum energy conformations of the dimeric subunit. Nucleic Acids Res. 1978 Sep;5(9):3249–3260. doi: 10.1093/nar/5.9.3249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hingerty B., Broyde S. Helix geometry of single stranded DNA 'A' and 'B' forms from minimum energy conformations of dimeric subunits. Nucleic Acids Res. 1978 Jan;5(1):127–137. doi: 10.1093/nar/5.1.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leslie A. G., Arnott S. Structure of the single-stranded polyribonucleotide poly(2'-O-methylcytidylic acid). J Mol Biol. 1978 Mar 5;119(3):399–414. doi: 10.1016/0022-2836(78)90222-x. [DOI] [PubMed] [Google Scholar]
- Neidle S., Taylor G., Sanderson M. A 1:2 crystalline complex of ApA:proflavine: a model for binding to single-stranded regions in RNA. Nucleic Acids Res. 1978 Nov;5(11):4417–4422. doi: 10.1093/nar/5.11.4417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olson W. K., Flory P. J. Spatial configuration of polynucleotide chains. II. Conformational energies and the average dimensions of polyribonucleotides. Biopolymers. 1972 Jan;11(1):25–56. doi: 10.1002/bip.1972.360110103. [DOI] [PubMed] [Google Scholar]
- Olson W. K., Flory P. J. Spatial configurations of polynucleotide chains. 3. Polydeoxyribonucleotides. Biopolymers. 1972 Jan;11(1):57–66. doi: 10.1002/bip.1972.360110104. [DOI] [PubMed] [Google Scholar]
- Olson W. K. The spatial configuration of ordered polynucleotide chains. I. Helix formation and base stacking. Biopolymers. 1976 May;15(5):859–878. doi: 10.1002/bip.1976.360150505. [DOI] [PubMed] [Google Scholar]
- Olson W. K. The spatial configuration of ordered polynucleotide chains. II. The poly(rA) helix. Nucleic Acids Res. 1975 Nov;2(11):2055–2068. doi: 10.1093/nar/2.11.2055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perahia D., Pullman B., Saran A. Molecular orbital calculations on the conformation of nucleic acids and their constituents. XI. The backbone structure of (3'-5') and (2'-5')-linked diribose monophosphates with different sugar puckers. Biochim Biophys Acta. 1974 Jun 14;353(1):16–27. doi: 10.1016/0005-2787(74)90093-8. [DOI] [PubMed] [Google Scholar]
- Perahia D., Pullman B., Vasilescu D., Cornillon R., Broch H. A molecular orbital investigation of the conformation of transfer RNA. Biochim Biophys Acta. 1977 Sep 20;478(2):244–259. doi: 10.1016/0005-2787(77)90188-5. [DOI] [PubMed] [Google Scholar]
- Renugopalakrishnan V., Lakshminarayanan A. V., Sasisekharan V. Stereochemistry of nucleic acids and polynucleotides. 3. Electronic charge distribution. Biopolymers. 1971;10(7):1159–1167. doi: 10.1002/bip.360100707. [DOI] [PubMed] [Google Scholar]
- Stellman S. D., Hingerty B., Broyde S. B., Subramanian E., Sato T., Langridge R. Structure of guanosine-3',5'-cytidine monophosphate. I. Semi-empirical potential energy calculations and model-building. Biopolymers. 1973 Dec;12(12):2731–2750. doi: 10.1002/bip.1973.360121208. [DOI] [PubMed] [Google Scholar]
- Tosi C., Clementi E., Matsuoka O. Conformational studies on polynucleotide chains. III. Intramolecular energy maps and comparison with experiments. Biopolymers. 1978 Jan;17(1):67–84. doi: 10.1002/bip.1978.360170106. [DOI] [PubMed] [Google Scholar]
- Viswamitra M. A., Kennard O., Jones P. G., Sheldrick G. M., Salisbury S., Favello L., Shakked Z. DNA double helical fragment at atomic resolution. Nature. 1978 Jun 22;273(5664):687–688. doi: 10.1038/273687a0. [DOI] [PubMed] [Google Scholar]
- Yathindra N., Sundaralingam M. Analysis of the possible helical structures of nucleic acids and polynucleotides. Application of (n-h) plots. Nucleic Acids Res. 1976 Mar;3(3):729–747. doi: 10.1093/nar/3.3.729. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yathindra N., Sundaralingam M. Backbone conformations in secondary and tertiary structural units of nucleic acids. Constraint in the phosphodiester conformation. Proc Natl Acad Sci U S A. 1974 Sep;71(9):3325–3328. doi: 10.1073/pnas.71.9.3325. [DOI] [PMC free article] [PubMed] [Google Scholar]


