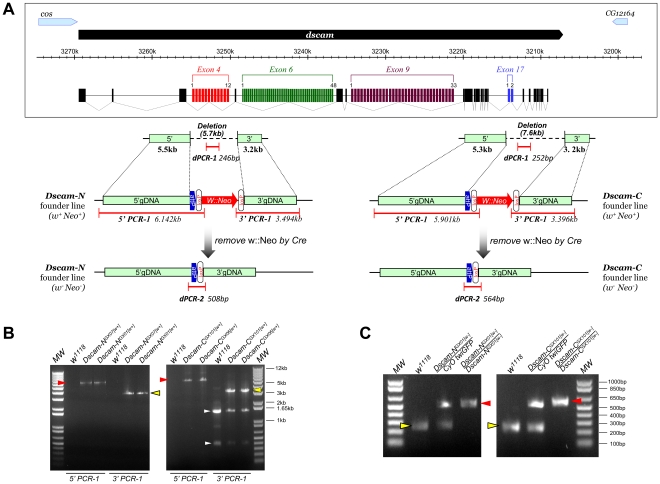
Figure 3. Gene targeting of Dscam-N and Dscam-C.
A. Targeting design and PCR verification of Dscam-N and Dscam-C founder lines. Boxed are the genomic DNA (gDNA) structure and alternative-splicing patterns of Dscam locus. Dscam locus contains four alternative-splicing exons: 4, 6, 9 and 17 [13]. Green boxes are gDNA regions used for 5′ and 3′ homology arms in the targeting constructs. In the Dscam-N founder knock-out line, a 5.7 kb genomic DNA covering the alternatively spliced exon 4 are deleted. In the Dscam-C founder knock-out line, a 7.6 kb genomic DNA covering the alternatively spliced exon 17 plus all the remaining downstream exons and 3′UTR are deleted. Dscam-N and Dscam-C founder knock-out lines carrying W::Neo marker are verified by 5′ and 3′ PCRs. 5′ or 3′ PCR is designed with one primer annealing within the W::Neo, while another primer anneals outside the gDNA region used for homology arms (“5′ gDNA” or “3′ gDNA”) in targeting constructs. Thus, only the correct targeting events will yield PCR products of expected size. Dscam-N and Dscam-C founder lines with W::Neo removed are further verified by dPCR-1 and dPCR-2. dPCR-1 is located within, while dPCR-2 spans over, the deleted region of Dscam-N or Dscam-C. B. 5′ and 3′ PCR-1 (red and yellow arrowheads, respectively) results from adults of Dscam-NGX07[w+]/CyO, Dscam-NGX01[w+]/CyO, Dscam-CGX101[w+]/CyO and Dscam-CGX06[w+]/CyO. w1118 was used as wild type control. White arrowheads pointing to non-specific PCR products. C. dPCR-1 (yellow arrowhead) and dPCR-2 (red arrowhead) results from embryos of Dscam-NGX01[w−] and Dscam-CGX101[w−] with W::Neo removed. Dscam-NGX01[w−] and Dscam-CGX101[w−] were balanced on CyO, twi-GAL4, UAS-2xEGFP (“CyO twiGFP”) chromosome so homozygous embryos could be distinguished by the absence of GFP. w1118 was used as the wild type control. For each PCR reaction genomic DNA was prepared by pooling approximately ten embryos together. For each sample, dPCR-1 and dPCR-2 reactions were carried out separately and were pooled before loading on the gel. MW: 1kb-plus DNA ladder (from Invitrogen); 5′ and 3′: the 5′ and 3′ homology arms of Dscam-N and Dscam-C targeting construct.